-
聚乙烯醇(PVA)通过聚醋酸乙烯酯水解制得,是一种具有良好性能且可生物降解的水溶性聚合物[1]。PVA因其良好的水溶性,粘附性,耐磨性和强韧性等优良的性能,被广泛用于农用地膜,纺织上浆和食品包装等行业[2]。PVA具有较大的表面活性,因此,大量降解缓慢的PVA广泛存在于自然界中,造成水体和泥土富氧、生态环境危害和生物链的破坏[1, 3]。许多研究报道了处理PVA的物理和化学的方法,如吸附法[4]、化学凝固法[5]、超声波降解法[6]、膜过滤法[7]和催化氧化法[8]。但这些方法成本较高且可能造成二次污染,而生物降解法更加高效和经济,因此,筛选高效降解菌和优化降解方式对于减少PVA污染而言具有重要意义[9]。
自1973年首株能够降解PVA的假单胞菌(Pseudomonas O-3)[10]被分离出以来,大量关于PVA降解菌的研究被报道。从染料厂废水中分离出的鞘氨醇盒菌(Sphingopyxis sp. PVA3)在6 d的培养中,对初始浓度为1 g·L−1 PVA的降解率达到90%[11]。粪产碱菌(Alcaligenes faecalis KK314)降解PVA的机理被详细报道,PVA上的羟基首先被氧化为β-羟基酮,然后被进一步水解为甲基酮类化合物和羧酸类化合物[12]。放线菌委内瑞拉链霉菌(Streptomyces venenzuelae GY1)能产生降解PVA的诱导型胞外酶[13]。地霉 (Geotrichum WF9101)对低分子质量的PVA具有降解的活性[14]。青霉菌(Penicillium sp. WSH02-21)能在12 d内将PVA降解完全,且能够分泌过氧化氢酶,防止氧化PVA过程中产生的过氧化氢对细胞产生损害[15]。共生降解也被广泛研究,其主要分为2种模式。首先是MORI等[16]报道的2种菌株共代谢,2株菌都不分泌生长因子。另外一种模式由SAKAZAWA等[17]提出。其中1株菌分泌生长因子,其他菌利用这种生长因子进行聚合物降解,如假单胞菌(Pseudomonas sp.VM15C)和恶臭假单胞菌(Pseudomonas putida VM15A)[18],新鞘氨醇菌(Novosphingobium sp.) 和黄色杆菌(Xanthobacter flavus)等[19]。由于PVA降解菌广泛存在于自然界[20],因此,进一步筛选高效菌并研究降解特性就具有独特意义。本研究通过对聚乙烯醇材料堆肥,研究了材料表面的生物多样性变化,并且从经历了3年降解的PVA材料表面筛选到了能独立降解PVA的苏云金芽孢杆菌(Bacillus thuringiensis sp.),命名为DG01。对DG01进行了摇瓶降解实验和呼吸降解实验,并对其降解特性进行了研究。
全文HTML
-
聚乙烯醇材料受赠于华南理工大学材料科学与工程学院;DNA提取试剂盒购于美国OMEGA公司;PVA2488(平均聚合度2 400±50,醇解度86.0%~90.0%)购于台湾长春化工有限公司。
-
将材料切割成若干长方块(长3.5 cm,宽 2.0 cm,高 1.0 cm)。将长方块置于超净台中紫外灭菌45 min。用商业型堆肥对材料进行覆盖,覆盖深度为10 cm。将被肥料覆盖好的材料置于常温下避光保藏。在降解过程中,适量加水与翻动土壤,保证堆肥的含氧量和湿度。在降解1、2、3 a时分别取样。每次取3个样,对材料表面细菌的多样性进行分析。
-
在聚乙烯醇材料降解过程中,用DNA提取试剂盒提取材料表面微生物的DNA。将目标时间段的材料置于装有50 mL无菌生理盐水的250 mL三角烧瓶中,适当振荡后,取出材料。按照试剂盒步骤提取DNA,提取液须−20 ℃保存。使用1%的琼脂糖进行电泳检查DNA样品是否合格。
-
选取通用引物(F338(5′-ACTCCTACGGGAGGCAGCA-3′)和R806(5′-GGACTACHVGGGTWTCTAAT-3′)),扩增细菌16S DNA片段的V3~V4区。PCR总体系为50 µL,包括25 µL premix,上下游引物各为0.5 µL,模板DNA 1 µL,ddH2O 23 µL。反应条件为:94 ℃预变性5 min;94 ℃变性30 s;58 ℃退火45 s;72 ℃延伸60 s,循环25次;最后72 ℃延伸10 min。样品储存的温度为4 ℃。用1.2%凝胶琼脂糖做凝胶电泳检测,最后由广州基迪奥生物科技有限公司对其测序。
-
在数据预处理时,基于序列末端相重合的部分,用FLASH将序列拼接起来。采用Usearch和Uchime除去非扩增区域序列和嵌合体。使用Prinseq 软件,控制过滤去除编码序列以及部分低质量序列。读段与接头序列对比超过15 bp长度,则认为接头污染。读段与接头序列超过9 bp长度,则认为质量较低。
在OTUs(operating taxonomic units)分类及Alpha多样性分析时,使用Mothur平台数据对tag进行去冗余处理,挑选出Unique tag。在相似度97%的水平上,对这些有效数据进行OTUs聚类分析、物种分析及稀释曲线分析。基于OTUs结果,用QIIME计算出多样性指数Chao、Ace和丰富度指数Shannon、Simpson。采用Bayesian算法,对代表OTUs进行分类并选取最佳分类水平,比较微生物多样性的变化。
-
选取经历了3年降解的材料,从材料表面筛选出了可降解PVA的菌株DG01。经过16S鉴定,株菌与Bacillus thuringiensis BM-BT15426(CP020723.1)相似度达到100%,命名为DG01。
-
LB培养基:蛋白胨 10.0 g·L−1,NaCl 5.0 g·L−1,葡萄糖 1.0 g·L−1,酵母膏粉 5.0 g·L−1,pH 7.0±0.2。
营养琼脂培养基:蛋白胨 10.0 g·L−1,牛肉浸出粉 3.0 g·L−1,氯化钠 14.0 g·L−1,琼脂 14.0 g·L−1,pH 7.0±0.2。
PVA无机盐培养基:PVA 24 883.0 g·L−1,酵母提取粉 1 g·L−1,NH4NO3 0.10 g·L−1,NaCl 0.02 g·L−1,MgSO4 0.05 g·L−1,CaCl2 0.05 g·L−1,FeSO4·7H2O 0.02 g·L−1,K2HPO4 1.60 g·L−1,KH2PO4 0.20 g·L−1,pH 7.0~7.2。
无机盐培养基:酵母提取粉 1.0 g·L−1,NH4NO3 0.10 g·L−1,NaCl 0.02 g·L−1,MgSO4 0.05 g·L−1,CaCl2 0.05 g·L−1,FeSO4·7H2O 0.02 g·L−1,K2HPO4 1.60 g·L−1,KH2PO4 0.20 g·L−1,pH 7.0~7.2。
-
从斜面上挑取适量菌株至100 mL(37 ℃,150 r·min−1)LB活化培养基中活化。活化16 h后,按无机盐培养基体积的5%接种菌液。
-
1)摇瓶降解实验。按培养方法,将接种了DG01的PVA无机盐培养基置于37 ℃,150 r·min−1 摇床中培养。每组3个平行,每隔48 h取样,测定培养基中PVA的浓度、pH、活菌浓度。
活菌浓度的测定:根据GB 4789.2-2016,对摇瓶降解实验的菌浓度进行测定。取1 mL的菌液,稀释至原菌液浓度的10−4、10−5、10−6倍;各取50 μL,涂布于营养琼脂培养基平板;将涂布好的平板置于37 ℃培养8 h左右,记录每个平板的菌落数;活菌浓度及其平均值按式(1)和式(2)计算。
式中:N为活菌浓度,CFU·mL−1;Ni为第i个样品的活菌浓度,CFU·mL−1;∑C为平板上的菌落之和;ni为稀释的倍数。
PVA浓度的测定:采用改良的finely法[21],取上清液,通过96孔板,测量PVA浓度和PVA降解率。取摇瓶培养后的发酵液,8 000g离心10 min后,保留上清液;将碘化钾-碘溶液60 μL(碘1.27 g·L−1、碘化钾2.5 g·L−1),上清液40 μL和100 μL硼酸(浓度4 g·L−1)依次加入96孔板;在避光4 ℃下反应15 min;用酶标仪检测波长为510 nm处的吸光度,通过标准曲线测定PVA的浓度,且以显色后的吸光度计算降解率。
降解率按照式(3)进行计算。
式中:η为PVA的降解率;A0为标准对照,降解前培养基的吸光度;A1为降解后培养基的吸光度;A为空白,无机盐培养基的吸光度。
-
2)以二氧化碳为指标的降解率的测定。根据EN ISO 14852-2004的方法搭建测定装置。在37 ℃下测试15 d。装置分为3部分:第1个部分吸收流入空气中的二氧化碳,由3个500 mL三角瓶组成,前2个瓶装200 mL的氢氧化钠溶液(0.05 mol·L−1,吸收二氧化碳),第3个瓶中装氢氧化钡溶液(0.012 5 mol·L−1),以检验二氧化碳是否除净;第2个部分由DG01利用PVA产生二氧化碳,由1个500 mL三角瓶组成,瓶子装有200 mL接种DG01后的PVA无机盐培养基;第3部分吸收由菌株产生的二氧化碳,由3个500 mL的三角烧瓶,分别装有200 mL 经过矫正的氢氧化钡溶液。气体流速控制在80 mL·min−1左右。用标准氢氧化钠(0.05 mol·L−1)矫正盐酸,然后用矫正后的盐酸滴定第3部分的氢氧化钡溶液来计算二氧化碳排放量。
以二氧化碳排放量为指标的降解率按照式(4)计算。
式中:η1为降解率;C0为装置第3部分氢氧化钡的浓度,mol·L−1;V0为装置中第3部分所含氢氧化钡的总体积,mL;C1为盐酸的浓度,mol·L−1;V1为滴定时消耗盐酸的总体积,mL;M为CO2的相对分子质量,g·mol−1;
${T_{{\rm{CO_2}}}} $ 为理论上排放的总二氧化碳,在本装置中取值为1.198 8 g。 -
3)DG01降解PVA2488最适条件研究。按培养方法将PVA无机盐培养基分别置于不同温度下(33、35、37、39、41、43、45 ℃),培养48 h,每组3个平行,测定降解前后的浓度,以此计算出PVA的降解率。考察最优降解温度。
按培养方法将DG01接种于酵母粉浓度不同(0.2、0.5、0.8、1.1、1.4、1.7、2.0 g·L−1)的无机盐培养基中,培养48 h,每组3个平行,测定降解前后PVA浓度,以此来计算PVA的降解率。考察最优酵母粉浓度。
配制初始pH不同的无机盐培养基(5.0、5.5、6.0、6.5、7.0、7.5、8.0)。按培养方法将DG01接种到配好的培养基中,培养48 h,每组3个平行,测定降解前后浓度,以此计算出PVA的降解率。考察最优初始pH。
1.1. 试剂与堆肥实验
1.2. 优势菌群分析
1.3. 菌种、培养基及培养方法
1.4. Bacillus thuringiensis DG01降解PVA2488实验
-
群落多样性分析:基于V3+V4区的测序,经过处理,最终从降解时间为1、2、3 a的材料表面的样品中分别获得427、556和604个OTUs。降解1、2、3 a样品的稀释曲线分别在600、550和400个左右趋于平缓,说明测序覆盖率良好,基本覆盖样品的全物种。由表1可以看出,降解2 a的Chao1指数达到最大值,Simpson指数微量增高。能利用聚乙烯醇材料生长的微生物渐渐汇集在材料表面,不能利用材料的微生物逐渐减少,导致多样性减少。堆肥带来大量的有机质以及营养物质,对汇集在材料表面的微生物有促进作用,使细菌的丰富度增加[22]。降解3 a的Chao1指数大幅度下降,Simpson大幅度增高。材料被长期覆盖后,其表面的结构,营养成分以及通氧量发生了改变,限制了菌株的生长,导致物种多样性和丰富度都下降[23]。
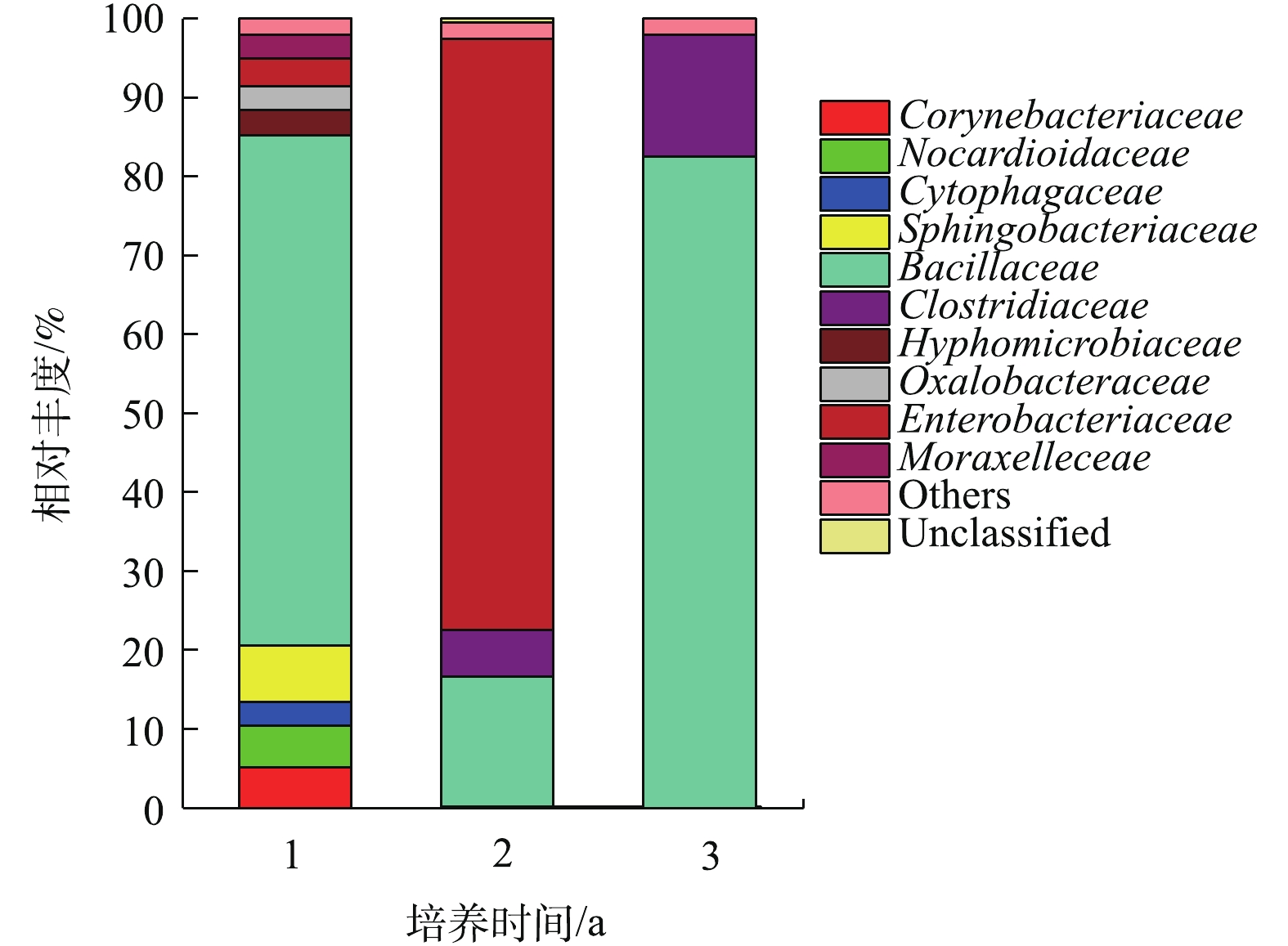
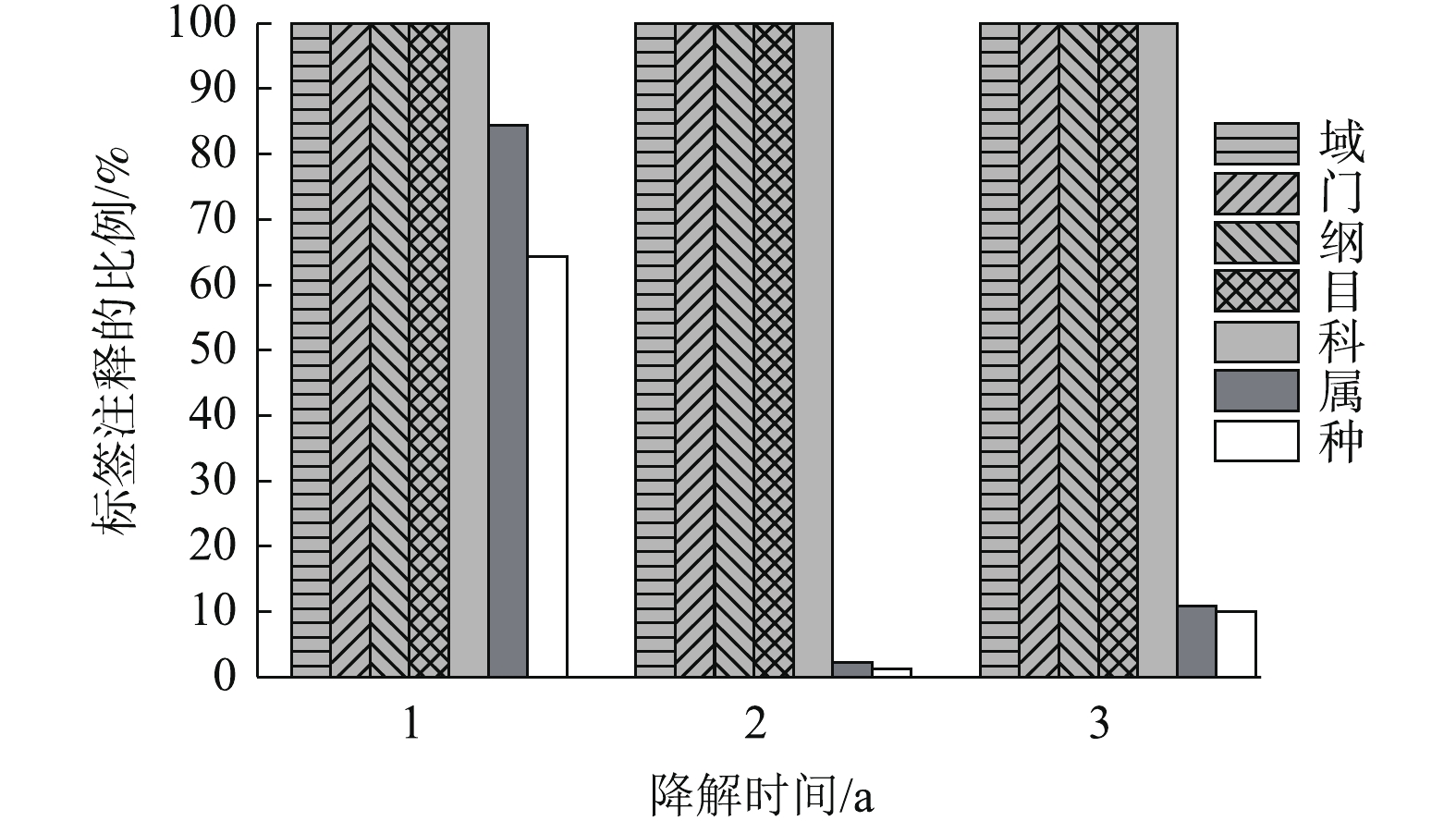
菌群结构变化及优势菌株分析:通过尽可能降低分类水平和提高tag的注释率,可获得最佳分类水平。分别在界、门、纲、目、科、属、种分类水平上统计tag序列数。结果如图1所示。在界、门、纲、目、科的分类水平上均有100%的tag序列能被注释。在分类水平属上,降解3 a的样品有接近80%的tag能被注释,降解1年和2年样品的tag只有10%能被注释。因此,科为最佳分类水平。
如图2所示,聚乙烯醇材料在堆肥中降解时,汇集在材料表面的菌群主要包括10个科(每个科的丰度在2%以上),分别是状杆菌科(Corynebacteriaceae)、诺卡氏菌科(Nocardioidaceae)、鞘氨醇杆菌科(Sphingobacteriaceae)、杆菌科(Bacillaceae)、细胞噬菌体科(Cytophagaceae)、梭状芽孢杆菌科(Clostridiaceae)、生丝微菌科(Hyphomicrobiaceae)、草杆菌科(Oxalobacteraceae)、肠杆菌科(Enterobacteriaceae)、摩拉克菌科(Moraxelleceae)。其中在3个时期都占主要成分是杆菌科(Bacillaceae),在降解的第1年和第3年都占绝大多数,相对丰度分别达到82.47%及64.62%。肠杆菌科(Enterobacteriaceae)主要在中前期被检测到,而在第3年几乎检测不到。梭状芽孢杆菌科(Clostridiaceae)随着时间增长其相对丰度也从5%缓慢增加到16%左右。其他7类菌株在前期丰度相对较少,在中后期几乎检测不出。在材料降解的第1年,第2年,第3年,Bacillaceae的丰度都明显较高,故认为其是优势菌。
-
从经历了3年降解的聚乙烯材料表面筛出了能降解聚乙烯醇的菌株。经16S rRNA分子生物学鉴定,该菌株属于在降解过程中占优势的杆菌科(Bacillaceae),命名为Bacillus thuringiensis DG01。能降解PVA的Bacillus thuringiensis鲜有报道。
-
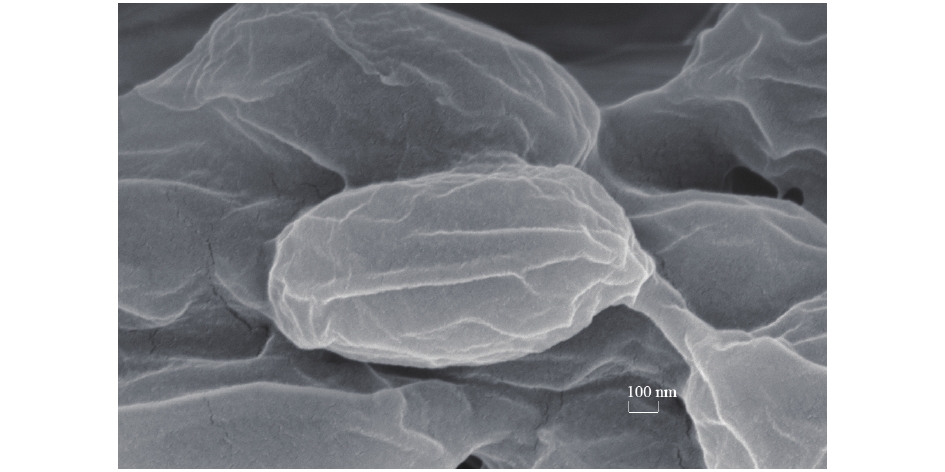
1)无机盐培养基下DG01形态SEM观察。对在PVA无机盐培养基里培养8 d的菌株进行电镜观察,结果如图3所示,DG01呈椭圆杆状长度和宽度各约为1.30 μm和0.64 μm。不同于在活化培养基的大小约1.4 μm×3.2 μm(图片没有给出),微生物形态变化可能与吸收PVA有关。HU等[24]发现,Spingopyxis sp.113P3在吸收PVA时,膜结构发生改变,以利于微生物吸收培养基中PVA。
-
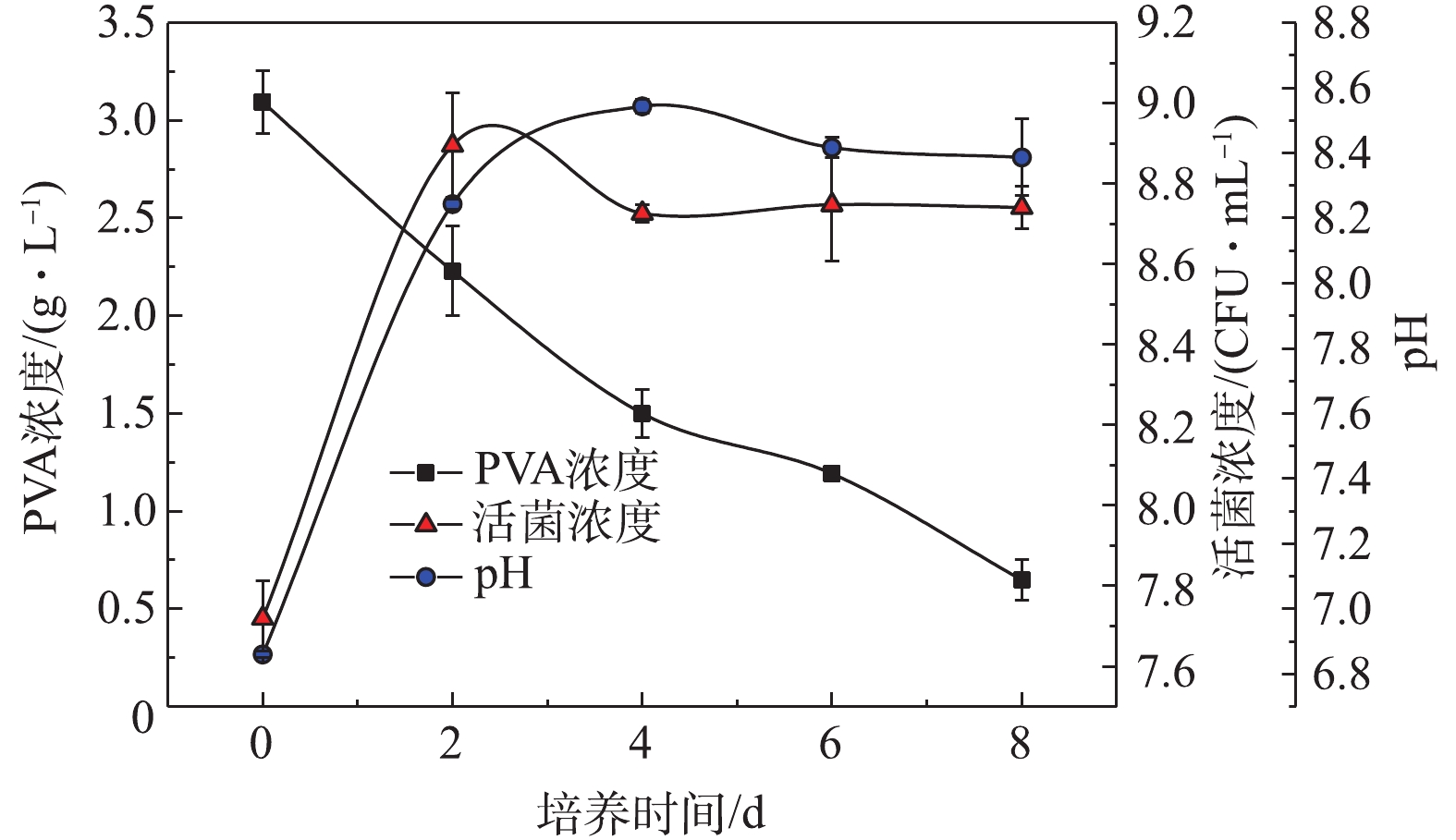
2) DG01降解PVA2488实验。Bacillus thuringiensis DG01降解PVA2488的过程如图4所示,活菌浓度在48 h内进入稳定期并达到最大值,在随后的6 d内维持不变。平衡期DG01的pH维持在8.20左右,降解环境维持稳定。对培养基中PVA浓度的下降趋势进行了动力学分析,结果表明PVA浓度下降趋势与一级动力学模型吻合良好,降解速率常数k和R2分别为0.195 d−1和0.984[25]。
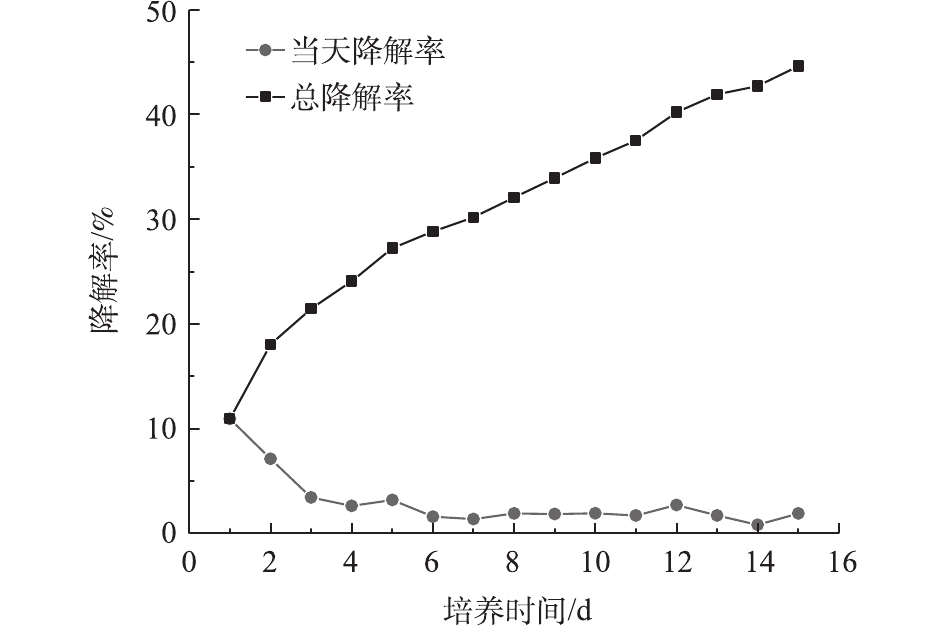
图5表明,DG01在前2 d降解率比较高,随后每天的降解率急剧下降。第1 天和第2 天的降解率之和达到18.04%,在后13 d内每天的降解率均较低。降解趋势符合一级动力学模型,R2为0.983 5。由图4和图5可知,以PVA浓度为指标表示的降解率与以二氧化碳排放量为指标表示的降解率不相符,后者的降解滞后于前者,说明细胞吸收的PVA并没有立即全部转化为CO2,降解时产生了中间体。目前普遍认为PVA的降解主要分为2步。首先,PVA上的羟基被氧化,形成单酮和β–二羰基结构;然后,这些单酮和β–二羰基被进一步水解,PVA分子链断开,生成羧酸和甲基酮类物质。通过重复以上2步反应,PVA分子链不断变短,形成短链羧酸类物质,这些物质最终进入微生物的中心代谢途径,成为微生物的能源物质和细胞生长可利用的碳源[3,26]。通过对比降解前后PVA2488的红外光谱图,发现有新的羧基产生。凝胶排阻色谱图表明降解前后PVA2488的分子质量有所减小。因此,认为在PVA2488在降解过程中,可能产生了分子链相对较短的羧酸类物质。
-
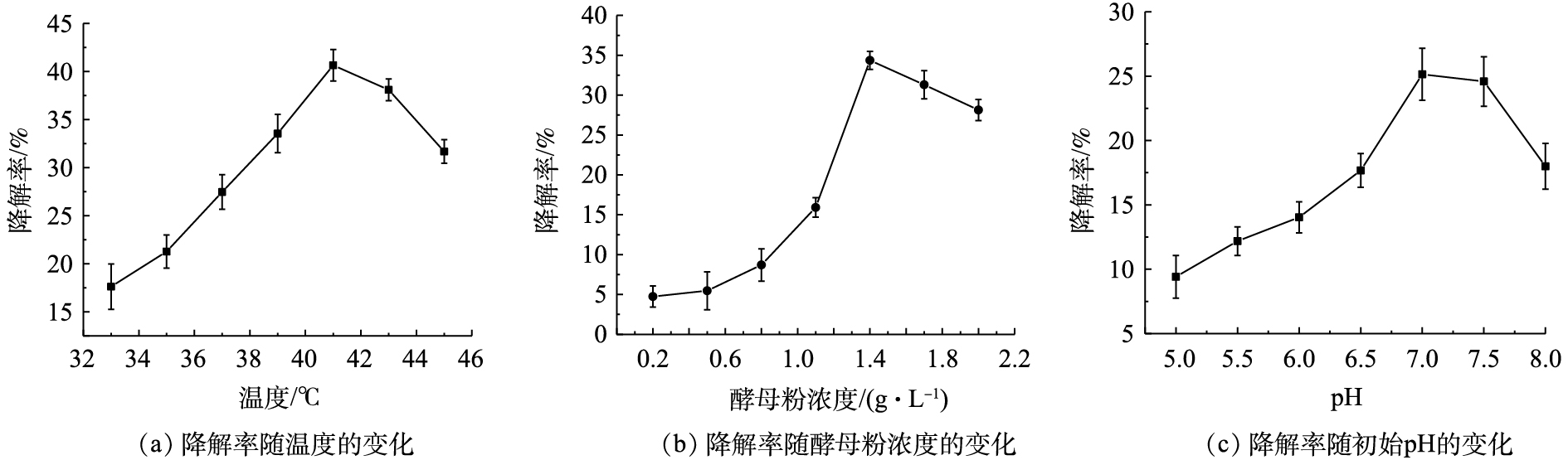
3) DG01降解PVA2488最适条件研究。分别对温度、初始pH和初始酵母粉浓度进行了单因素优化,结果如图6所示。当培养温度为41 ℃ 时,PVA2488降解率最高,可达到40.65%;当初始酵母粉浓度为1.4 g·L−1时,PVA2488降解率最高,可达到34.36%;当初始pH为7.0时,PVA2488降解率最高,可达到25.14%。因此,选择最优培养温度为41 ℃,最优初始酵母粉浓度为1.4 g·L−1,最优初始pH为7.0。
按培养方法,在酵母粉浓度为1.4 g·L−1,初始pH为7.0的PVA培养基中接种DG01。在温度为41 ℃的条件下培养48 h,测量3个平行样降解前后培养基中PVA浓度,计算其降解率。结果表明,在最适条件下,DG01对初始浓度为3 g·L−1 PVA的降解率可达到45.21%,降解了1.356 g·L−1的PVA,降解率提高了2.10倍。优化后PVA降解速率为0.678 g·(L·d)−1,高于Stenotrophomonas rhizophila QL-P4[26]和Sphingopyxis sp.PVA3[11],其降解速率在PVA初始浓度为1 g·L−1时分别为0.350 g·(L·d)−1和0.125 g·(L·d)−1。也高于Pseudomonas sp.strain VM15C[18]和Penicillium sp.WSH02-21[15], 其降解速率在PVA初始浓度为5 g·L−1时分别为0.500 g·(L·d)−1和0.417 g·(L·d)−1。由此可见,DG01有相对较高的降解速率,有潜在的应用前景。
2.1. 聚乙烯醇材料表面的菌群分析
2.2. Bcaillus thuringiensis DG01降解特性实验
-
1) 通过高通量测序,分析了聚乙烯醇复合材料降解过程中材料表面种群结构的变化,并找到了优势种群Bacillaceae,为堆肥降解PVA的研究提供了基础。在接下来的研究中将对堆肥中降解PVA菌群的功能进行分析。
2) 从降解了3 a的材料中筛选出属于优势种群Bacillaceae的菌株Bacillus thuringiensis DG01。分别以培养基中PVA浓度和CO2排放量为指标研究降解趋势。2种指标下的降解趋势都符合一次降解动力学模型,且培养基中被微生物吸收的PVA没有完全转化为CO2。进一步研究发现,PVA在降解过程中产生了羧酸类物质,为研究该菌株降解PVA的机理做出了指导。在接下来的研究中将进一步详细探讨该菌株降解PVA的过程。
3) 对DG01降解PVA2488的条件进行了优化,结果表明,其最适温度、初始pH和酵母粉浓度分别为41 ℃、7.0和1.4 g·L−1。在最佳条件下,3 g·L−1 的PVA在24 h的降解率为45.21%,提高了2.10倍。为该菌株降解PVA的实际应用打下了坚实基础。



 下载:
下载: