-
规律间隔成簇短回文重复序列及其相关蛋白(clustered regularly interspaced short palindromic repeats/CRISPR associated proteins,CRISPR/Cas)系统是古细菌和大多数细菌在生物进化过程中抵御病毒入侵而形成的一种适应性免疫防御系统. 1987年Nakata等[1]首次报道了它的特殊结构,crRNA(CRISPR RNA)包含间隔序列与重复序列[2],间隔序列来源于噬菌体和共轭质粒等外源基因,用于识别目标核酸,重复序列则用于结合Cas蛋白并指引其发挥作用. 随后科学家们在超过40%的细菌和90%的古生菌中发现了类似结构的序列[3],命名为CRISPR/Cas[4]. 对Cas蛋白的研究发现Cas12a和Cas13a不仅可以特异性顺式切割目标双链DNA(dsDNA)、单链DNA/RNA(ssDNA/RNA),还具有非特异性切割溶液中的游离核酸的“反式切割”[5 − 7]. 由于CRISPR/Cas系统具有特异性识别目标核酸、非特异性切割游离核酸及可编程性等优点,无需专业设备即可提供价格低廉、快速、准确的即时检测,被用来开发快速、高效、低成本和高灵敏度的分子传感器,例如SHERLOCK[8]和DETECTR[9]等,在新型冠状病毒检测中发挥了重要作用[10 − 12].
近年来,除重金属离子外,抗生素、内分泌干扰物、持久性难降解有机物等新型污染物越来越受人们关注,已成为环境分析的重要靶物. 传统上,这些非核酸靶标的检测依靠高效液相色谱(HPLC)[13]、气相色谱/质谱(GC/MS)[14]或酶联免疫吸附测定(ELISA)[15]等方法. 然而,这些方法通常耗时长、成本高,或需要昂贵的仪器和专业操作人员,不利于现场即时检测或快速筛查. 鉴于CRISPR/Cas系统的成熟可靠和可扩展性,研究人员尝试将其应用于环境/食品安全领域的非核酸靶标检测中,开发出了一系列高灵敏度、高特异性的便携式检测设备. 然而,相对于核酸检测应用,CRISPR/Cas系统在非核酸靶标的检测研究相对滞后. 这主要因为Cas的活化依赖于核酸底物,而构筑高灵敏度、高特异性和通用性的元件将非核酸靶标信息转换为核酸信息仍然面临挑战. 为此,本文综述了基于CRISPR/Cas系统的生物传感器在环境监测领域中非核酸靶标检测的应用进展,包括了重金属离子、阴离子、新污染物、农药、真菌毒素以及有害细菌等目标物,重点讨论了将非核酸靶标的信息转换为核酸信息的策略,并对不同的信号输出方式和检测能力进行分析比较(信息汇总于表1),最后对该领域研究存在的问题和可能的发展方向进行探讨.
-
重金属离子对环境的污染可直接或间接对人体造成严重危害. 传统的重金属检测依赖于大型检测仪器,成本高且耗时长,因此建立一种方便、高效、灵敏的检测方法很有必要. 借助于可特异性识别重金属离子的RNA-cleaving DNAzyme(RCD)、适配体以及变构转录因子等作为信息转换元件,CRISPR系统已被用来构建快速检测重金属离子的生物传感器. 这些方法具有很高的灵敏度、操作简便、成本低以及可扩展等优点,具有应用前景. 本节重点介绍利用以上信息转换元件和CRISPR系统构筑Pb2+[16 − 20]、Cd2+[21]、Zn2+[22]等几种重金属离子的生物传感器以及应用潜力,并对存在的问题进行探讨.
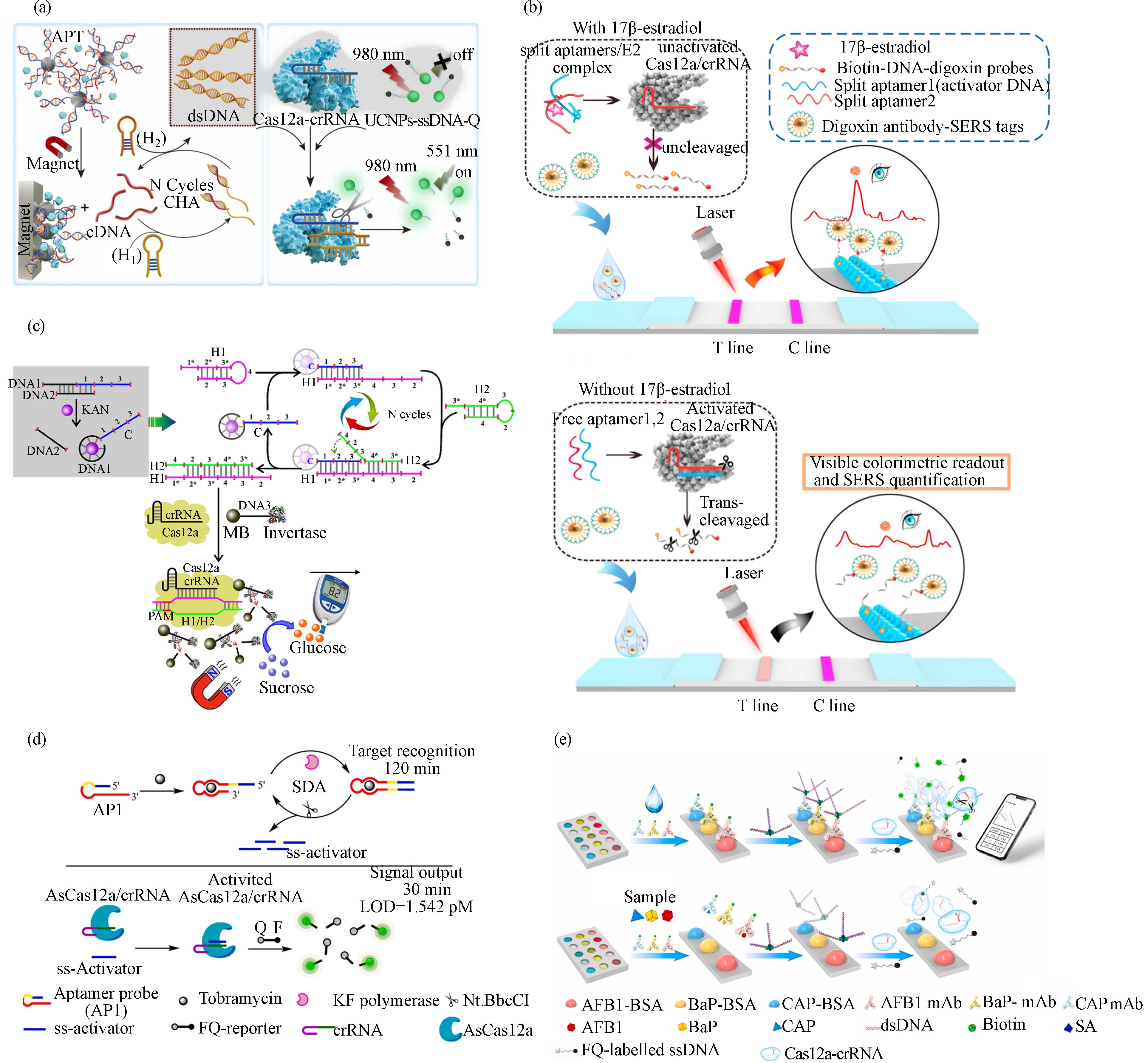
RCD是通过体外筛选技术(in vitro selection)得到的具有催化功能的DNA序列,可以在特定辅因子的作用下切割对应RNA底物. 目前,已有不少针对特定金属离子的RCD被公开报道[44 − 45]. RCD切割底物释放的片段可被CRISPR/Cas系统识别,从而使CRISPR/Cas的核酸切割活性被激活,该过程可将对金属离子的识别转换为核酸切割信号. 该策略仅需在RCD的底物一端设计可被crRNA特异性识别的DNA序列,在特定重金属离子作用下RCD将底物切割为两段;将可被crRNA识别的片段转移到CRISPR/Cas系统中,从而激活Cas蛋白切割报告分子. 上述CRISPR/Cas系统还可与第二个RCD组合,通过切割报告分子产生的片段再激活此RCD,形成多酶串联的信号放大系统,可以方便地通过荧光、比色、电化学等方式进行输出. 基于上述原理,科学家构建了基于RCD-CRISPR/Cas系统的检测Pb2+的生物传感器[16]. 首先在底物的5’端设计可被crRNA特异性识别的目标链,并在底物3’端标记生物素使其被固定在链霉亲和素磁珠上. 当Pb2+存在时,RCD将底物切割为两段,磁吸移除带有生物素的一段产物,然后将含目标链的产物转移到CISPR/Cas系统,与crRNA识别激活Cas蛋白. 最后,Cas蛋白切割标记有荧光和猝灭基团报告分子,从而产生信号. 经过优化研究,发现底物长度增加到20-nt时,切割效率最佳,检出限(LOD)达到0.053 nmol·L−1,远低于饮用水中铅的浓度限值(72 nmol·L−1). 然而,由于RCD的底物常含有rA碱基,在检测实际样品如血清样品时,存在被核酸酶降解产生较高背景信号的问题. 有研究报道在高盐条件下,寡核苷酸在纳米颗粒上密集排列成三维球形,具有很强的稳定性和抗降解能力[46 − 47]. 因此,采取将RCD的底物固定在金纳米颗粒(AuNP)形成球形核酸的方法,有助于提高其稳定性,降低背景信号. 该策略组合CRISPR/Cas12a来检测Pb2+(图1a)[17],最低检测限可以达到fmol·L−1级别,并且在实际样本如血清、空气颗粒和土壤中也有很好的检测性能. 该系统还可与芯片检测设备整合,用智能手机显示检测结果,提高现场检测的实用性.
利用荧光报告分子可以快速、准确地显示结果,但荧光修饰的报告分子价格昂贵,而且依赖荧光检测设备. 为降低成本,开发了基于CRISPR/Cas12a-RCD的比色传感器[18]. 在该平台中,将报告分子设计为MnO2NRs(氧化锰纳米棒)-DNA-MB(磁珠)复合材料,当存在目标分析物时,报告分子断裂,携带MnO2NRs的产物片段氧化TMB产生肉眼可见的颜色变化. 该系统被用于酒和食用油等样本中Pb2+的检测,回收率达到97%—104%,LOD达到0.54 nmol·L−1. 虽然其灵敏度相比荧光法有所下降,但可视化的信号输出方式使其在实验室外的检测更为便捷.
环境中重金属离子的浓度通常较低,对传感器的灵敏度的要求高. 基于CRISPR/Cas的传感器检测核酸分析物时,可将目标核酸进行预扩增处理,如滚环扩增(Rolling Circle Amplification, RCA)、链置换扩增技术(Strand Displacement Amplification, SDA)、催化发夹自组装(Catalytic Hairpin Assembly, CHA)及杂交链式反应(Hybridization Chain Reaction, HCR)等,以提升灵敏度. 这些放大策略同样也可和CRISPR/Cas整合来检测Pb2+[19]. 首先目标分析物激活RCD切割底物,随后掉落的产物底物片段启动SDA扩增,产生可被crRNA识别的DNA,进而引导Cas蛋白切割报告分子. 该平台的信号输出采用电化学设备,将报告分子ssDNA-Fc(二茂铁)在ZrO2/CeO/PAH的帮助下固定在玻璃碳电极上,根据报告分子断裂引起电流的变化来判断是否存在目标物. 该方法对Pb2+的检测限达到0.02 pmol·L−1,可检测酒、油、大米等样本中的Pb2+.
适配体(aptamer)和变构转录因子(allosteric transcription factor, aTF)也被用作CRISPR/Cas系统的信息转换元件. 研究人员在CRISPR/Cas12a中引入了Cd2+的适配体,构建了可检测Cd2+的生物传感器(图1b)[21]. 在SDA的引物P设计了Cd2+的适配体序列,当样品中没有Cd2+时,引物与模板结合后在DNA聚合酶的作用下产生dsDNA. 随后,Nt.BbvCI切口酶在引物延伸区域生成聚合酶的识别位点,启动二次延伸,同时产生大量的ssDNA被crRNA特异性识别并引导Cas蛋白切割荧光报告分子. 当存在Cd2+时,Cd2+与适配体结合,阻止引物P与模板T的结合,使SDA的效率降低,荧光信号变弱. 该方法检测Cd2+的LOD可达到60 pmol·L−1,远低于美国环保署饮用水中Cd2+的限量标准(44.5 nmol·L−1),回收率为97.59%—104.94%,具有很好的实际应用潜力. 研究人员还利用响应Zn2+的aTF作为信息转换元件构建了检测Zn2+的CRISPR/Ca13a生物传感器[22]. 目标分析物的存在使aTF与DNA的亲和力减弱,从而转录出可被crRNA特异性识别的激活剂,引导Cas蛋白酶对底物进行反式切割,产生可被检测的荧光信号. 在市政水样中,Zn2+浓度为1 μmol·L−1时可以利用手持式荧光检测设备观察到荧光信号.
总的来说,在重金属离子检测中,RCD仍然是CRISPR/Cas系统的主要信息转换元件. 目前,依赖Cu2+、Ag+、Cr3+、Uo2+以及镧系金属离子Lu3+等具有生态环境风险的重金属离子发挥作用的RCD已经被报道[48 − 50],这些进展为CRISPR/Cas系统在环境分析中的应用提供了新机遇. 然而,不少RCD对金属离子的专一性较差[44],因此仍有必要优化筛选条件,获得更具选择性的识别工具. 另外,高特异性区分Fe2+和Fe3+的RCD已经被筛选出来[51],启示如果能够筛选依赖不同价态(如Cr3+和Cr6+)或不同形态(如有机汞和无机汞)的RCD可能具有实际应用价值. 目前已报道的RCD的催化转化速率大多低于蛋白质酶(目前公认活性最高的RCD为10-23 DNAzyme,kcat ≈ 10 min−1)[44],而且实际应用中RCD的催化活性易受如pH、温度、有机溶剂等条件的影响[52]. 因此,开发能够耐受多种环境条件、催化活性更高的RCD,将有助于RCD和CRISPR/Cas系统整合的生物传感器的实际应用. 除RCD以外,能够专一性识别某些重金属离子如Zn2+、Cd2+、Pb2+、Co2+、Hg2+和Cu2+的适配体也被陆续报道[53 − 58],这为基于CRISPR的金属离子传感器的信息转换元件提供了更多选择.
-
水体中富含诸多无机阴离子,如F−、Cl−、NO2−、Br−、NO3−、PO43−、SO32−、SO42−等,当其含量超过一定阈值,可能具有潜在的生态风险. 响应氟离子(F−)的核糖开关作为转导元件与CRISPR/Cas系统整合,在大肠杆菌内构建了F−传感器(图1c)[22]. 在该系统中,RNA聚合酶的识别位点被F−核糖开关占据. 当F−与核糖开关结合时,RNA聚合酶可执行转录产生RNA,然后被crRNA特异性识别,引导Cas蛋白酶反式切割报告分子,产生荧光信号. 该系统对F−的LOD可达1 μmol·L−1. 然而,由于大肠杆菌的RNA对氟化物极为敏感,F−浓度超过100 μmol·L−1时抑制Cas的蛋白活性,从而限制了其线性范围. 为了准确的定量氟化物的浓度,研究人员将细胞内的转录过程移到体外,构建了无细胞的F−传感器,获得了更宽的线性响应范围[23]. 在该系统中,DNA模板包含启动子、间隔序列、核糖开关以及靶向crRNA的序列. 当F−存在时,F−核糖开关直接触发转录产生RNA,被crRNA特异性识别激活Cas蛋白切割报告分子,该方法使F−的检测上限提高到800 μmol·L−1,并且LOD为1.7 μmol·L−1,可检测瓶装水、自来水和湖水中的F−. 然而,除F−外,对于Br−、NO3−、SO42−等众多可溶于水的阴离子,仍缺乏特异性强、亲和力高的信息转换元件,限制了CRISPR/Cas系统在阴离子检测中的应用.
-
新污染物如持久性有机污染物、内分泌干扰物、药品和个人护理品等对于生态环境和人体健康构成潜在威胁,已经成为国际上高度关注的环境问题[59]. 目前,新污染物的检测主要依赖于可精确分析多种污染物分子的大型仪器,对于污染物类型的环境调查或非靶向监测具有重要作用. 然而,对于具有重要风险的污染物的靶向监测,可能更需要能在现场应用的快速检测工具,而这些大型仪器则难以适应.
核酸适配体是指通过指数富集的配体系统进化技术(SELEX)筛选得到的一段寡核苷酸序列. 核酸适体可折叠形成特定三维结构,以高亲和力和特异性结合靶标,其靶标范围涵盖金属离子、小分子、蛋白质到细胞等多种类型[60]. 相较于抗体,适配体易合成、易修饰、稳定性强、靶分子范围广,因此在环境分析领域也有诸多应用[61 − 62]. 理论上,适配体与靶分子结合引起的构象变化[63 − 65],可产生被CRISPR/Cas系统识别的核酸激活剂. 得益于筛选技术的快速发展,研究人员已经获得了不少对新污染物类分子具有高亲和力和选择性的适配体,使得适配体作为CRISPR/Cas系统检测这些化合物的信息转换元件具有应用潜力[60,66]. 此外,小分子的抗体也可作为CRISPR/Cas系统的信息转换元件,研究人员通常将抗体与可被crRNA识别的DNA以生物素和链霉亲和素连接,进而通过抗体-抗原的特异性识别将非核酸靶转换为核酸信号.
-
内分泌干扰物也被称为环境激素,是一大类具有内分泌干扰毒性的物质. 常见的内分泌干扰物有在塑料中大量使用的双酚A(BPA)、邻苯二甲酸酯类化合物以及天然和人工合成的类固醇激素等化合物. 这些物质在自然环境和人类生活中无处不在,对动物和人类的生殖、发育等产生持久性危害,导致多种与内分泌相关的疾病,严重威胁生态系统和人类健康. 雌二醇(E2)和BPA是广受关注的内分泌干扰物,二者的适配体作为信息转换元件被整合进CRISPR/Cas系统,构建了生物传感器[24]. 作者首先通过适配体链将DNA激活剂锁住,当目标分析物存在时,与适配体结合引起其构象变化,释放激活剂DNA片段,从而被crRNA识别激活Cas蛋白. 该方法对E2和BPA检测限分别为0.08 nmol·L−1和0.06 nmol·L−1,并且在污水和尿液中仍具有很好的分辨能力. 为了拓宽E2检测的应用场景,可视化的信号输出方式被引入适配体-CRISPR/Cas12a的生物传感器[25]. 该策略通过适配体将非核酸靶标转化为核酸信号,并通过CHA来放大信号,扩增产物被crRNA识别并引导Cas蛋白将DNA水凝胶包被的金属有机框架(MOF)裂解,催化TMB与 H2O2反应产生颜色变化(图2a). 该系统采用上转换材料替代荧光团,有效地提高了信噪比和荧光稳定性,并通过CHA将信号进一步放大,提高了检测的灵敏度. 并通过牛奶、猪肉等样品验证了其实际应用能力,与荧光法相比,该方法的灵敏度略低(LOD = 0.015 ng·mL−1).
与传统的信号输出方式相比,侧向层析测定(Lateral flow assay, LFA)因其操作简便、分析时间短、价格低廉备受关注. Li团队[26]将LFA用于信号输出,开发了一种检测E2的CRISPR/Cas-适配体生物传感器. 该平台主要由三部分组成(图2b):报告分子为Biotin-ssDNA-抗地高辛抗体复合物,LFA的测试区涂抹链霉亲和素,控制区涂抹抗小鼠IgG抗体,以及用于信号显示的地高辛抗体-Au@BDT@Au复合物. E2与适配体结合可使Cas蛋白失去活性,不切割报告分子;测试区的链霉亲和素捕获Biotin标记的抗地高辛抗体以及用于信号输出的地高辛抗体-Au@BDT@Au,产生信号,剩余样品流至控制区. 相反,不存在E2时,适配体序列被crRNA特异性识别,引导Cas蛋白切割报告分子,导致地高辛抗体-Au@BDT@Au不能被捕获在测试区,因而测试区域信号降低或没有信号. 该方法结合了CRISPR/Cas12a的信号放大能力和间隙增强拉曼标签(Au@BDT@Au)的高灵敏度,较传统的LFA灵敏度有很大提升(LOD = 180 fmol·L−1),可用于实际样品包括血清、尿液、牛奶和自来水中的E2检测.
-
抗生素广泛用于人类医学和畜牧业,然而抗生素的滥用及其在水环境和土壤中的累积间接地危害人类健康,在临床中常用的抗生素包括磺胺、喹诺酮、β内酰胺、四环素、大环内酯、氨基糖苷等六大类. 目前,已报道了基于CRISPR系统开发的卡那霉素[27 − 29]、妥布霉素[30]、四环素[31]、氨苄青霉素[32 − 33]等的生物传感器,在抗生素的快速现场检测中具有应用前景. 基于适配体-CRISPR/Cas检测非核酸靶的策略大多通过一条适配体链将激活链锁住,这种方法可能会产生较高的背景值. 为降低背景信号,研究人员使用两段卡那霉素的适配体序列将激活剂锁定[27]. 该系统的报告分子铥(Tm)-Rep-DNA-Bio(生物素)被裂解后,携带生物素的部分被链霉亲和素磁珠分离,带有Tm-Rep的一段可以通过同位素法监测. 该方法可以在30 min内检测卡那霉素,LOD低至4.06 pmol·L−1. 该方法使用镧系元素Tm,虽然在生物和环境中的干扰极低,但应用成本高,对仪器设备要求高. 为此,研究人员开发了成本更低、灵敏度更高的适配体-CRISPR/Cas12a系统[28]. 该系统仍采用适配体与DNA链互锁的方式,通过加入靶分子诱导适配体构象变化,使被其锁住的DNA链释放,启动HCR,生成被crRNA特异性识别的dsDNA,引导Cas蛋白切割报告分子. 报告分子ssDNA两端分别标记磁珠和葡萄糖转化酶,被切断后,分离带有转化酶的片段,催化蔗糖水解成葡萄糖,最后用便携式血糖仪定量葡萄糖来评估卡那霉素的浓度(图2c). 该方法对卡那霉素的LOD达到了1 pmol·L−1. 此外,适配体-CRISPR/Cas系统的信号也可被LFA输出[29],检测卡那霉素的LOD达到了14.8 nmol·L−1,该系统用于牛奶中卡那霉素检测的效果与市售试纸条相当.
Li等[30]在CRISPR/Cas系统中引入适配体构建了两种用于妥布霉素检测的生物传感器sensor-ss和sensor-ds. 在sensor-ss中,采用ssDNA作为激活剂并经过SDA扩增以提高传感器的灵敏度. 当目标分析物存在时,适配体结构发生变化使得激活剂被释放并启动SDA,扩增产物被crRNA识别引导Cas蛋白切割荧光报告分子(图2d). 而sensor-ds的激活剂则是使用含有原间隔序列邻近基序位点(PAM)的dsDNA,未采用核酸扩增技术放大信号. 结果显示sensor-ss(LOD = 1.542 pmol·L−1)比sensor-ds(LOD = 3.719 pmol·L−1)具有更高的灵敏度,但由于经过扩增导致反应过快达到检测平台其检测线性范围较窄(前者5—30 pmol·L−1,后者10—300 pmol·L−1).
-
持久性有机污染物的种类很多,具有环境持久性、生物累积性、远距离迁移性和高毒性等特点. 此类污染物分子苯环多,如多环芳烃、多氯联苯、多溴联苯醚等,分子结构相似且水溶性很差,筛选核酸适体或制备抗体的难度大,限制了CRISPR系统的应用. 目前开发的基于CRISPR/Cas的工具仅用于少数分子的检测. 研究人员利用抗体-抗原的特异性识别实现信息转换,与CRISPR/Cas系统整合(称为“iPOCT”)用于苯并[a]芘(Benzo (a) pyrene, BaP)的检测[34]. 该检测体系包括:固定在微孔板由小分子偶联牛血清白蛋白(BSA)构建的抗原,生物素化的抗体,链霉亲和素标记的dsDNA—用来结合crRNA并激活Cas蛋白,标记荧光和淬灭基团的ssDNA报告分子(图2e). 当待测样品中没有目标分子时,固定化的抗原与生物素化的抗体结合,然后捕获链霉亲和素标记的dsDNA;洗去未结合的dsDNA,再向微孔板内加入CRISPR/Cas系统,保留在孔内的dsDNA结合crRNA,激活Cas蛋白切割报告分子产生荧光增强的信号. 待测样品中含有目标分子,则游离的待测分子与生物素化的抗体结合,导致抗体与固定化的抗原的结合受到抑制,从而使微孔捕获的dsDNA含量减少,进而减少了CRISPR/Cas切割底物的荧光增强信号. 除了BaP,该策略也成功用于其他小分子化合物的检测,说明该策略具有一定的通用性,而且还可通过酶标仪进行高通量检测多个目标化合物. 优化后的传感器对目标分子的LOD达到4.971 fg·mL−1,在湖水等实际样品中也有很好的检测能力. 然而,该策略检测目标分子时输出的信号低于非目标物对照的信号,属于信号减小的响应模式. 此外,基于CRISPR生物传感器也被用来检测与持久性有机污染物类似的对羟基苯甲酸分子 [35].
虽然基于CRISPR系统的生物传感器在新污染物检测中展示了应用前景,但受限于信息转换元件的可及性,此类生物传感器尚未得到普遍应用. 以抗体作为靶分子识别与信息转换元件的CRISPR系统在新污染物检测中的应用较少,主要在于新污染物大多为分子量小于1000Da的小分子,没有免疫原性,很难制备相应的抗体. 适配体因其广泛的分子识别能力,作为CRISPR的信息转换元件更有优势. 目前已有一些新污染物分子,如全氟辛酸(PFOA)[67]、多氯联苯(PCB77)[68]、邻苯二甲酸酯(PAEs)[69]、四环素类、氨基糖苷类、β-内酰胺类、大环内酯类、氯霉素类、喹诺酮类等抗生素[70]的适配体被筛选出,为基于CRISPR的检测技术开发奠定了基础. 然而,新污染物的种类很多,很多分子具有相似的结构且水溶性差,缺少活性位点,使得通过靶分子固定的方法筛选适配体挑战性很大,所得适配体对结构非常相似的分子的选择性不高. 以上因素制约了适配体作为CRISPR的信息转换元件在新污染物检测中的实际应用. 发展新的筛选技术,获得亲和力更高、选择性更好、能够适应实际检测环境的适配体,将有助于推动CRISPR系统在新污染物检测中的实际应用.
-
虽然RCD是基于CRISPR/Cas系统的生物传感器的理想信号转换元件,但目前尚未有专门响应某一农药分子的RCD被开发出来,限制了该元件在农药分子检测的直接使用. 为此,研究人员设计了乙酰胆碱酯酶-RCD-CRISPR/Cas三酶级联的策略来检测有机磷农药(图3a)[36]. 首先,有机磷农药的加入使乙酰胆碱酯酶水解乙酰硫代胆碱产生硫代胆碱,硫代胆碱将MnO2纳米片还原成Mn2+,激活依赖Mn2+的RCD将底物切断释放激活剂,最后激活剂被crRNA特异性识别并引导Cas蛋白切割报告分子,产生荧光信号. 该策略成功检测了实际样品柑橘类水果、卷心菜中的对氧磷、敌敌畏和内吸磷的3种农药,LOD分别达到了270、406、218 pg·mL−1,远低于农药使用标准中的最低残留量. 然而,有机磷农药对乙酰胆碱酯酶的活性影响没有特异性,应用该方法不能区分具体的农药分子,在实际应用中仅可用于该类农药的初筛. 为实现特异性检测某一农药成分,研究人员将适配体引入CRISPR/Cas系统构建了检测啶虫脒的传感器[37]. 首先将啶虫脒适配体组装成球形核酸,将对啶虫脒的识别信号转换为核酸信号,进而激活Cas蛋白的活性,可在生菜中检测该农药残留检测(LOD = 2.7 pmol·L−1),为农药残留监测提供了更加精准、便捷的方法.
-
真菌毒素是真菌在农作物、食品中产生的次级代谢产物,可在人体中累积对人体健康造成危害. 常见的真菌毒素有黄曲毒素类、赭曲毒素A及镰孢菌毒素类,此类毒素具有很强的毒性以及致癌性,是环境监测真菌毒素中的重点关注对象. 利用抗体作为CRISPR/Cas系统的信号转换元件的策略被用来开发脱氧雪腐镰刀菌烯醇(deoxynivalenol, DON)的传感器[38]. 作者首先制作了DON抗体用以捕获目标分析物,其次通过ssDNA将PS微球与磁性纳米颗粒(MNPs)连接构建信号探针,最后是将DON通过牛血清白蛋白(BSA)与激活crRNA的DNA链偶联. DON的存在会使抗体捕获DON-激活剂,从而被crRNA特异性识别引导Cas蛋白切割报告分子PS-ssDNA-MNP,然后通过磁吸将PS微球分离出去,通过粒子计数器测量PS微球的数量用于DON的定量分析. 该方法对DON的LOD 达到了0.061 ng·mL−1,与传统的ELISA和HPLC相比表现出更宽的线性范围和更高的灵敏度. 此策略也用于黄曲霉毒素B1(AFB1)检测[34]. 然而,这些毒素的分子量小于
1000 kD,没有免疫原性,通常需要将它们制成半抗原偶联到蛋白质分子如BSA上,再进行免疫反应制备抗体,不仅流程繁琐而且难以保证选择性;另外抗体稳定性较差,保存困难. 适配体被称为化学抗体,针对小分子的核酸适体可通过SELEX技术筛选得到. 目前科学家已经分离了不少特异性结合AFB1的适配体,其结构也得到了详细表征[71],为开发基于适配体的AFB1检测工具打下坚实基础. 研究人员将AFB1的适配体与CRISPR/Cas系统联用构建检测AFBI的平台(图3b)[39],利用crRNA锁住适配体,当靶标存在时引起适配体构象变化,抑制其与crRNA的结合,降低Cas蛋白的切割能力. 该方法在酒、牛奶样品中实现了AFB1的高灵敏度检测(LOD = 0.8 ng·mL−1). 该系统的关键在于crRNA要与适配体-靶标结合的区域互补配对,这样靶标的加入才会对crRNA和适配体杂交产生很大影响,为此作者设计了28种crRNA用来优化与适配体杂交的位点. 该研究提示如果适配体与靶分子相互作用的机制明确,将对设计适配体-CRRISPR/Cas的分子传感器很有帮助.鉴于单一信号输出方式常常限制了检测范围或LOD,因此提出了基于CRISPR/Cas-适配体的双信号生物传感器[40]. 首先将报告分子ssDNA-Fc固定在金电极表面,ssDNA-Fc通过碱基互补将适配体固定,而适配体通过与激活剂1的结合互相锁定. 加入胶霉毒素时,诱导适配体构象变化,释放激活剂1,使其与H4ETTC-ssDNA结合产生聚集诱导发光;H4ETTC-ssDNA锁定的激活剂2则被释放,与crRNA识别,引导Cas切割ssDNA-Fc产生电化学信号(图3c). 传感器通过电化学、荧光双信号输出,具有很高的灵敏度(LOD = 2.4 fmol·L−1),作者应用该系统对苹果、玉米等食物样品中胶霉毒素残留进行检测. 借鉴上转换材料相较于荧光探针的优势,有研究人员利用其构建了CRISPR生物传感器用于赭曲霉毒素的监测[41].
农药和真菌毒素的残留监测对于环境/食品安全具有重要意义,目前科学家已开发出了多个针对农药和真菌毒素的适配体[72 − 73],这为基于CRISPR/Cas检测农药、真菌毒素研究提供了强有力的支持. 然而,利用适配体作为信息转换元件常常需要靶分子诱导其发生构象变化,因此需要研究人员更系统地研究适配体与靶分子的相互作用机制,以提高分子设计的成功率.
-
有害细菌的快速检测也是环境监测和食品安全检测的重要任务. 与细菌培养、抗体分析和核酸分析相比,CRISPR/Cas系统为病原菌的灵敏检测提供了一种简单便捷的方法. 研究人员将适配体引入CRISPR/Cas系统,并结合HCR设计了一种检测鼠伤寒沙门氏菌的生物传感器[42](图4a). 当靶标存在时,竞争结合适配体以启动HCR,HCR的产物可以被crRNA特异性识别并引导Cas蛋白切割报告分子. 通过HCR扩增,显著增强了反式切割活性,从而实现了灵敏检测(LOD = 20 CFU·mL−1),并可用于牛奶样品中的鼠伤寒沙门氏菌检测,提高了在临床诊断的应用潜力.
目前基于CRISPR/Cas的检测方法都使用线性crRNA(LcrRNA)作为引导RNA. 然而,在自然界中存在一种共价闭合环状RNA,可以通过化学或酶连接的方法制备,目前很少被用于CRISPR生物传感系统. 鉴于此,Wu等[43]将crRNA设计成环状引导RNA的形式(CcrRNA),证实了环化会使crRNA失去引导作用,无法激活正常的Cas蛋白的切割反应. 随后,研究团队使用大肠杆菌依赖性DNAzyme和肺炎克雷伯菌依赖的RCD作为信息转换元件,用于识别大肠杆菌和肺炎球菌. 当目标分析物存在时,DNAzyme活性被激活,切割CcrRNA形成LcrRNA(如图4b). 这样就使得crRNA获得引导作用,引导Cas蛋白切割报告分子. 经过优化,使得大肠杆菌和肺炎球菌的LOD达到了102 CFU·mL−1,并使用该系统对尿路感染的患者进行诊断,灵敏度和特异性分别达到100%、90%. 该方法将环化RNA设计进CRISPR-Cas系统,使CRISPR Cas系统摆脱线性crRNA的束缚,极大降低了背景值的同时,还灵活应用了细菌特异性的RCD(也称为适配体酶)切割RNA的特性. 此外利用RCD切割环化crRNA可以在同一管内实现信号转换和Cas蛋白的激活,避免了繁琐的实验步骤. 这也是首次通过控制crRNA的结构变化来调节CRISPR反应活性,为后续开发RCD耦联CRISPR/Cas系统用于非核酸靶标检测提供了新思路.
-
基于CRISPR/Cas的生物传感系统具有高特异性、可编程性及高催化活性,在快速、灵敏、特异的核酸检测中极具应用前景. 然而,在非核酸靶标的研究方面,该系统尚处于起步阶段. 由于Cas蛋白的活化依赖于核酸底物,因此将非核酸识别信息转换为核酸信息是其应用的关键. 目前的信息转换策略中,以DNAzyme和适配体作为信息转换元件较为普遍. 这些策略在非核酸靶标的检测上取得了良好的开端,但其进一步应用仍面临着诸多挑战.
基于CRISPR/Cas系统的非核酸靶标检测依赖于信号转导元件. 虽然已有研究通过逻辑门的方法实现了多目标分析物的检测,然而受信息转换元件的限制,其应用范围有限,尚未见类似SHERLOCK[8]这样通用的平台被开发出来. 近期科学家开发了一种将小型化和自组装的微阵列系统与基于CRISPR的特异性核酸检测相结合的方法[74],建立了一种可扩展的多重检测平台,这为基于CRISPR/Cas系统的非核酸靶标的高通量检测提供了思路.
相对于环境监测的目标物而言,已开发并实际应用的转换元件仍然很少. 而且,现有的转换元件,如针对金属离子的DNAzyme,针对小分子的适配体,还存在特异性不够强的问题. 因此筛选更多、特异性更好的分子识别工具,是未来对环境分析样品实现即时现场快速检测的重要内容. 再者,适配体用于分子识别一般不能直接输出信号,大多经过复杂的分子设计实现构象转换等来释放信号,除了设计非常复杂,还可能导致亲和力下降. 这些因素制约了适配体与CRISPR/Cas系统的整合应用. Li课题组[75 − 76]以及Liu课题组[77]筛选得到的特异性识别有害细菌的DNAzyme,在进行分子识别的同时输出信号,是典型的智能分子. 这种系统可以方便地与CRISPR/Cas整合,这对推动CRISPR/Cas系统在环境分析中的应用有重要价值.
在环境分析中,目标分析物通常以较低浓度存在,这对传感器的灵敏度提出了挑战. 基于电化学法的信号输出具有很高的灵敏度,然而其抗干扰能力可能较差,应用于复杂环境中的分析物监测存在挑战. 开发抗污的电化学分析材料或设备,对于应用该系统进行环境分析有重要价值.
CRISPR/Cas系统的大多数识别或信息转换元件都涉及核酸,这些分子多在水溶液中开发. 然而,实际的环境分析对象,如新污染物,大多难溶于水,需要有机溶剂提取和溶解,因此开发适用于含有机溶剂的体系的分子识别/信息转换工具箱具有应用价值. 我们团队首先在含35% DMSO的缓冲溶液体系中筛选鉴定了一种高度依赖该条件的DNAzyme[78],并构建了一种能在35% DMSO体系中对靶分子进行识别并产生核酸切割信号的适配体酶系统,证明了可以在含高浓度有机溶剂的环境中筛选出高活性的DNAzyme. 为开发难溶于水且需要有机溶剂溶解的有害物质的快速检测工具提供了新思路,也提示有必要在真实的环境样品或应用场景中筛选出具有专一性强的DNAzyme、适配体或具有定向进化适应能力强的CRISPR/Cas系统,以提升生物传感器在环境分析的实际应用价值.
基于CRISPR/Cas系统的生物传感器在环境分析中的应用进展
Advances in CRISPR/Cas-based biosensors for environmental analysis applications
-
摘要: CRISPR/Cas可改造为靶核酸刺激-响应的传感系统,可视为集“分子识别与信号输出”一体的生物传感器,目前在核酸即时诊断中已得到广泛应用. 鉴于该系统具有成熟、可靠和可扩展的优点,研究人员尝试将其用于环境分析领域的非核酸分析物检测,已开发了一系列具有应用前景的便携式设备. 然而,如何将非核酸靶标信息转换为核酸信息仍存在挑战,这限制了CRISPR/Cas系统在环境分析中的应用推广. 为此,本文综述了CRISPR/Cas系统在重金属离子和阴离子、新污染物、农药与真菌毒素以及有害细菌等非核酸靶标检测中的应用进展,重点介绍了构筑非核酸靶标信息转换元件的策略,以期为推进该系统在环境分析中的应用提供支持.
-
关键词:
- CRISPR/Cas /
- 新污染物 /
- 核酸适体 /
- RNA-cleaving DNAzyme /
- 生物传感器.
Abstract: The CRISPR/Cas can be engineered as a stimuli-responsive sensing system for the target nucleic acids, which enables it to serve as a “two-in-one” biosensor with both molecular recognition element and signal output element. Up to date, this type of biosensing systems have been widely employed in the Point of Care Testing (POCT) for nucleic acids. Taking advantage of being mature, reliable and scalable, the CRISPR/Cas system has been developed as portable devices for the identification of non-nucleic acid analytes in the field of environmental analytical chemistry. However, challenges remain to be resolved in converting the information from non-nucleic acid targets to nucleic acids, and thus limiting its utilization in environmental analysis. Here, we review the applications of the CRISPR/Cas system in the detection of non-nucleic acid targets, including heavy metal ions and anions, emerging contaminants, pesticides, mycotoxins, and harmful bacteria. Moreover, we focus on the strategies of constructing non-nucleic acid target transducers in order to promote the utilization of this system in environmental analysis.-
Key words:
- CRISPR/Cas /
- emerging contaminants /
- aptamer /
- RNA-cleaving DNAzyme /
- biosensor.
-

-
图 1 基于CRISPR/Cas系统的离子传感器:(a)RCD-AuNP球形核酸与CRISPR/Cas整合的Pb2+传感器[17];(b)适配体-CRISPR/Cas系统检测Cd2+[21];(c)核糖开关-CRISPR/Cas系统检测F−[22]
Figure 1. CRISPR/Cas-based biosensor for the detection of heavy metal ions and anion (F−): (a) RCD-AuNP spherical nucleic acids and CRISPR/Cas system for the sensing of Pb2+ [17]; (b) An aptamer-CRISPR/Cas system as a Cd2+ sensor [21]; (c) A riboswitch-CRISPR/Cas system for the detection of F− [22]
图 2 基于CRISPR/Cas系统的内分泌干扰物、抗生素和持久性有机污染物的生物传感器:(a)适配体-CRISPR/Cas系统检测E2[25];(b)整合ELISA与CRISPR/Ca系统检测E2[26];(c)基于适配体-HCR-CRISPR/Cas的卡那霉素传感器[28];(d)基于适配体-SDA-CRISPR/Cas的妥布霉素传感器[30];(e)检测BaP、AFB1和CAP的“iPOCT”系统[34]
Figure 2. CRISPR/Cas-based biosensors for the detection of endocrine disrupting chemicals, antibiotics, and persistent organic pollutants: (a) Aptamer-CRISPR/Cas system for the sensing of E2[25]; (b) Combination of ELISA and CRISPR/Cas system for the detection of E2[26]; (c) Aptamers, HCR, and CRISPR/Cas-based biosensors for kanamycin[28]; (d) Aptamer-SDA-CRISPR/Cas system for Tobramycin detection[30]; (e) An “iPOCT” system for the detection of BaP, AFB1, and CAP[34]
图 3 基于CRISPR/Cas系统检测农药和真菌毒素的生物传感器:(a)检测农药的三酶级联系统 [36];(b)适配体-CRISPR/Cas系统检测AFB1[39];(c)适配体-CRISPR/Cas系统检测胶霉毒素[40]
Figure 3. CRISPR/Cas -based biosensors for the detection of pesticides and mycotoxins: (a) A three-enzymes-cascade system for the detection of pesticides[36]; (b), and (c) Aptamer-CRISPR/Cas-based biosensor for AFB1 (b) [39]; and gliotoxin (c) [40]
图 4 基于CRISPR/Cas检测细菌的生物传感器. (a)适配体-CRISPR/Cas系统检测鼠伤寒沙门菌[42];(b)基于RNA-cleaving DNAzyme切割ccrRNA激活CRISPR/Cas系统的策略检测大肠杆菌和肺炎克雷伯菌[43]
Figure 4. CRISPR/Cas-based biosensors for the detection of bacteria: (a) Aptamer-CRISPR/Cas-based biosensor for Salmonella typhimurium[42]; (b) A CRISPR/Cas-based biosensor that is activated by a ccrRNA-cleaving DNAzyme for the detection of Escherichia coli and Klebsiella pneumoniae[43]
表 1 基于CRISPR/Cas系统的传感器在环境分析中的应用
Table 1. Application of CRISPR/Cas-based biosensors in the environmental analysis field
靶标
Targets信息转
换元件
Information converting element效应蛋白
The type of Cas检出限
Limit of detection检测时间
Detection
time信号输出
Signal output样品
Samples检测设备
Testing equipment文献
Ref.重金属离子 Pb2+ DNAzyme(GR-5) Cas12a 0.053 nmol·L−1 15 min 荧光法 — — [16] Pb2+ DNAzyme(8-17E) Cas12a 86 fmol·L−1 — 荧光法 血清、空气颗粒、土壤 智能手机 [17] Pb2+ DNAzyme(GR-5) Cas12a 0.54 nmol·L−1 — 比色法 食用油、白酒 肉眼观察 [18] Pb2+ DNAzyme(GR-5) Cas12a 0.02 pmol·L−1 — 电化学 酒、花生、大米、食用油 电化学分析仪 [19] Pb2+ DNAzyme(GR-5) Cas12a 0.48 nmol·L−1 — 荧光法 矿泉水 便携式3D打印设备 [20] Cd2+ 适配体 Cas12a 60 pmol·L−1 120 min 荧光法 湖水、大米 — [21] Zn2+ 变构转录因子(SmtB) Cas13a 1 μmol·L−1 20 min 荧光法 市政水样 便携式3D打印设备 [22] 阴离子 F− 核糖开关(crcB) Cas13a 1 μmol·L−1 20 min 荧光法 — 便携式3D打印设备 [22] F− 核糖开关 Cas13a 1.7 μmol·L−1 30 min 荧光法 瓶装水、自来水、湖水 荧光计 [23] 内分泌
干扰物双酚A、
雌二醇适配体 Cas12a 0.06、
0.08 nmol·L−120 min 荧光法 尿液、污水 — [24] 雌二醇 适配体 Cas12a 0.015 ng·mL−1 — 比色法 牛奶、蛋类、猪肉 肉眼观察 [25] 内分泌
干扰物雌二醇 适配体 Cas12a 180 fmol·L−1 — 比色法 牛奶、自来水、血清、尿液 LFA [26] 抗生素 卡那霉素 适配体 Cas12a 4.06 pmol·L−1 30 min 同位素法 野生鱼血清、肌肉、肝脏 电感耦合等离子体-质谱法 [27] 卡那霉素 适配体 Cas12a 1 pmol·L−1 — 酶催化蔗糖转化为葡萄糖 河水、牛奶 血糖仪 [28] 卡那霉素 适配体 Cas12a 14.8 nmol·L−1 26min 比色法 牛奶 侧向层析测定 [29] 妥布霉素 适配体 Cas12a 1.542 pmol·L−1 — 荧光法 湖水、牛奶 紫外灯 [30] 四环素 变构转录因子(TetR) Cas12a 2 μmol·L−1 — 荧光法 环境水样 手持可视化荧光仪 [31] 氨苄青霉素 适配体 Cas12a 0.01 nmol·L−1 30 min 荧光法 鲜奶、生蛋清、生蜂蜜 荧光仪 [32] 氨苄青霉素 适配体 Cas14a 2.06 nmol·L−1 45 min 同位素法 江河水样 ICP-MS [33] 持久性有机污染物 苯并[a]芘 抗体 Cas12a 4.971 fg·mL−1 — 荧光法 湖水、大豆油 智能手机 [34] 对羟基
苯甲酸变构转录因子 Cas12a 1.8 nmol·L−1 — 荧光法 — 荧光仪 [35] 农药 对氧磷、
敌敌畏、
内吸磷DNAzyme
(8-17E变体)Cas12a 270、406、
218 pg·mL−1— 荧光法 柑橘类水果、卷心菜 — [36] 啶虫脒 适配体 Cas12a 2.7 pmol·L−1 — 电化学发光 生菜 肉眼观察 [37] 真菌毒素 脱氧雪腐镰刀菌烯醇 抗体 Cas12a 0.061 ng·mL−1 30 min 粒子计数 玉米、水 粒子计数器 [38] 黄曲霉毒素 抗体 Cas12a 0.00257 fg·mL−1— 荧光法 花生、面粉 智能手机 [34] 黄曲霉毒素 适配体 Cas12a 0.8 ng·mL−1 20 min 比色法 红酒、啤酒、牛奶 荧光板读取仪 [39] 胶霉毒素 适配体 Cas12a 2.4 fmol·L−1 55 min 电化学 苹果、胡萝卜、红薯、马铃薯、玉米 手持式电化学分析仪 [40] 赭曲霉毒素 适配体 Cas12a 0.83 ng/mL 60 min 荧光法 玉米粉 荧光仪 [41] 细菌 沙门氏菌 适配体 Cas12a 20 CFU·mL−1 — 电化学 牛奶 电化学工作站 [42] 大肠杆菌 DNAzyme
(EC1)Cas12a 102 CFU·mL−1 107 min 荧光法 尿液 荧光仪 [43] 肺炎球菌 DNAzyme
(KP6)Cas12a 102 CFU·mL−1 — 荧光法 — 荧光仪 [43] “—”,文献缺乏相应信息. “—”, Lack of information. -
[1] ISHINO Y, SHINAGAWA H, MAKINO K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product[J]. Journal of Bacteriology, 1987, 169(12): 5429-5433. doi: 10.1128/jb.169.12.5429-5433.1987 [2] MOJICA F J, DIEZ-VILLASENOR C, GARCIA-MARTINEZ J, et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements[J]. Journal Of Molecular Evolution, 2005, 60(2): 174-182. doi: 10.1007/s00239-004-0046-3 [3] MOJICA F J, DIEZ-VILLASENOR C, SORIA E, et al. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria[J]. Molecular Microbiology, 2000, 36(1): 244-246. doi: 10.1046/j.1365-2958.2000.01838.x [4] JANSEN R, EMBDEN J D, GAASTRA W, et al. Identification of genes that are associated with DNA repeats in prokaryotes[J]. Molecular Microbiology, 2002, 43(6): 1565-1575. doi: 10.1046/j.1365-2958.2002.02839.x [5] QIN J J, WANG W, GAO L Q, et al. Emerging biosensing and transducing techniques for potential applications in point-of-care diagnostics[J]. Chemical Science, 2022, 13(10): 2857-2876. doi: 10.1039/D1SC06269G [6] KIM S, JI S, KOH H R. CRISPR as a Diagnostic Tool[J]. Biomolecules, 2021, 11(8): 1162. doi: 10.3390/biom11081162 [7] YUE H H, HUANG M Q, TIAN T, et al. Advances in clustered, regularly interspaced short palindromic repeats (CRISPR)-based diagnostic assays assisted by micro/nanotechnologies[J]. ACS Nano, 2021, 15(5): 7848-7859. doi: 10.1021/acsnano.1c02372 [8] GOOTENBERG J S, ABUDAYYEH O O, LEE J W, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2[J]. Science, 2017, 356(6336): 438-442. doi: 10.1126/science.aam9321 [9] CHEN J S, MA E, HARRINGTON L B, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity[J]. Science, 2018, 360(6387): 436-439. doi: 10.1126/science.aar6245 [10] BROUGHTON J P, DENG X D, YU G X, et al. CRISPR–Cas12-based detection of SARS-CoV-2[J]. Nature Biotechnology, 2020, 38(7): 870-874. doi: 10.1038/s41587-020-0513-4 [11] SELVAM K, NAJIB M A, KHALID M F, et al. RT-LAMP CRISPR-Cas12/13-Based SARS-CoV-2 Detection Methods[J]. Diagnostics, 2021, 11(9): 1646. doi: 10.3390/diagnostics11091646 [12] LIANG Y H, LIN H Q, ZOU L R, et al. CRISPR-Cas12a-Based Detection for the Major SARS-CoV-2 Variants of Concern[J]. Microbiology Spectrum, 2021, 9(3): e0101721. doi: 10.1128/Spectrum.01017-21 [13] RANJBARIAN F, SHARMA S, FALAPPA G, et al. Isocratic HPLC analysis for the simultaneous determination of dNTPs, rNTPs and ADP in biological samples[J]. Nucleic Acids Research, 2022, 50(3): e18. doi: 10.1093/nar/gkab1117 [14] ZHANG S M, WANG H B, ZHU M J. A sensitive GC/MS detection method for analyzing microbial metabolites short chain fatty acids in fecal and serum samples[J]. Talanta, 2019, 196: 249-254. doi: 10.1016/j.talanta.2018.12.049 [15] ALBERTI D, VAN'T ERVE M, STEFANIA R, et al. A quantitative relaxometric version of the ELISA test for the measurement of cell surface biomarkers[J]. Angewandte Chemie International Edition, 2014, 53(13): 3488-3491. doi: 10.1002/anie.201310959 [16] LI J J, YANG S S, ZUO C, et al. Applying CRISPR-Cas12a as a Signal Amplifier to Construct Biosensors for Non-DNA Targets in Ultralow Concentrations[J]. ACS Sensors, 2020, 5(4): 970-977. doi: 10.1021/acssensors.9b02305 [17] LI Y Y, LI H D, FANG W K, et al. Amplification of the Fluorescence Signal with Clustered Regularly Interspaced Short Palindromic Repeats-Cas12a Based on Au Nanoparticle-DNAzyme Probe and On-Site Detection of Pb2+ Via the Photonic Crystal Chip[J]. ACS Sensors, 2022, 7(5): 1572-1580. doi: 10.1021/acssensors.2c00516 [18] XU S Q, WANG S T, GUO L, et al. Nanozyme-catalysed CRISPR-Cas12a system for the preamplification-free colorimetric detection of lead ion[J]. Analytica Chimica Acta, 2023, 1243: 340827. doi: 10.1016/j.aca.2023.340827 [19] YUE Y Y, WANG S T, JIN Q, et al. A triple amplification strategy using GR-5 DNAzyme as a signal medium for ultrasensitive detection of trace Pb2+ based on CRISPR/Cas12a empowered electrochemical biosensor[J]. Analytica Chimica Acta, 2023, 1263: 341241. doi: 10.1016/j.aca.2023.341241 [20] CHEN Y J, WU H, QIAN S W J, et al. Applying CRISPR/Cas system as a signal enhancer for DNAzyme-based lead ion detection[J]. Analytica Chimica Acta, 2022, 1192: 339356. doi: 10.1016/j.aca.2021.339356 [21] MA X C, SUO T Y, ZHAO F R, et al. Integrating CRISPR/Cas12a with strand displacement amplification for the ultrasensitive aptasensing of cadmium(II)[J]. Analytical and Bioanalytical Chemistry, 2023, 415(12): 2281-2289. doi: 10.1007/s00216-023-04650-6 [22] IWASAKI R S, BATEY R T. SPRINT: a Cas13a-based platform for detection of small molecules[J]. Nucleic Acids Research, 2020, 48(17): e101. doi: 10.1093/nar/gkaa673 [23] MA Y, MOU Q B, YAN P, et al. A highly sensitive and selective fluoride sensor based on a riboswitch-regulated transcription coupled with CRISPR-Cas13a tandem reaction[J]. Chemical Science, 2021, 12(35): 11740-11747. doi: 10.1039/D1SC03508H [24] ZHAO Y Q, ZHU L, DING Y X, et al. Simple and cheap CRISPR/Cas12a biosensor based on plug-and-play of DNA aptamers for the detection of endocrine-disrupting compounds[J]. Talanta, 2023, 263: 124761. doi: 10.1016/j.talanta.2023.124761 [25] WANG Y, PENG Y, LI S, et al. The development of a fluorescence/colorimetric biosensor based on the cleavage activity of CRISPR-Cas12a for the detection of non-nucleic acid targets[J]. Journal of Hazardous Materials, 2023, 449: 131044. doi: 10.1016/j.jhazmat.2023.131044 [26] LI Q, LI X B, ZHOU P X, et al. Split aptamer regulated CRISPR/Cas12a biosensor for 17beta-estradiol through a gap-enhanced Raman tags based lateral flow strategy[J]. Biosensors and Bioelectrons, 2022, 215: 114548. doi: 10.1016/j.bios.2022.114548 [27] HU J Y, SONG H J, ZHOU J, et al. Metal-Tagged CRISPR/Cas12a Bioassay Enables Ultrasensitive and Highly Selective Evaluation of Kanamycin Bioaccumulation in Fish Samples[J]. Analytical Chemistry, 2021, 93(42): 14214-14222. doi: 10.1021/acs.analchem.1c03094 [28] CHEN J H, SHI G, YAN C. Portable biosensor for on-site detection of kanamycin in water samples based on CRISPR-Cas12a and an off-the-shelf glucometer[J]. Science of The Total Environment, 2023, 872: 162279. doi: 10.1016/j.scitotenv.2023.162279 [29] LI X P, CHEN X J, MAO M X, et al. Accelerated CRISPR/Cas12a-based small molecule detection using bivalent aptamer[J]. Biosensors and Bioelectronics, 2022, 217: 114725. doi: 10.1016/j.bios.2022.114725 [30] LI D W, LING S, WU H S, et al. CRISPR/Cas12a-based biosensors for ultrasensitive tobramycin detection with single- and double-stranded DNA activators[J]. Sensors and Actuators B: Chemical, 2022, 355: 131329. doi: 10.1016/j.snb.2021.131329 [31] MAHAS A, WANG Q C, MARSIC T, et al. Development of Cas12a-Based Cell-Free Small-Molecule Biosensors via Allosteric Regulation of CRISPR Array Expression[J]. Analytical Chemistry, 2022, 94(11): 4617-4626. doi: 10.1021/acs.analchem.1c04332 [32] YEE B J, SHAFIQAH N F, MOHD-NAIM N F, et al. A CRISPR/Cas12a-based fluorescence aptasensor for the rapid and sensitive detection of ampicillin[J]. International Journal of Biological Macromolecules, 2023, 242(Pt 4): 125211. [33] HU J Y, ZHOU J, LIU R, et al. Element probe based CRISPR/Cas14 bioassay for non-nucleic-acid targets[J]. Chemical Communications, 2021, 57(80): 10423-10426. doi: 10.1039/D1CC03992J [34] ZHAO Y, WU W Q, TANG X Q, et al. A universal CRISPR/Cas12a-powered intelligent point-of-care testing platform for multiple small molecules in the healthcare, environment, and food[J]. Biosensors and Bioelectronics, 2023, 225: 115102. doi: 10.1016/j.bios.2023.115102 [35] LIANG M D, LI Z L, WANG W S, et al. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules[J]. Nature Communications, 2019, 10(1): 3672. doi: 10.1038/s41467-019-11648-1 [36] FU R J, WANG Y W, LIU Y L, et al. CRISPR-Cas12a based fluorescence assay for organophosphorus pesticides in agricultural products[J]. Food Chemistry, 2022, 387: 132919. doi: 10.1016/j.foodchem.2022.132919 [37] LI Y, YANG F, YUAN R, et al. Electrochemiluminescence covalent organic framework coupling with CRISPR/Cas12a-mediated biosensor for pesticide residue detection[J]. Food Chemistry, 2022, 389: 133049. doi: 10.1016/j.foodchem.2022.133049 [38] LI L T, HONG F, PAN S X, et al. "Lollipop" particle counting immunoassay based on antigen-powered CRISPR-Cas12a dual signal amplification for the sensitive detection of deoxynivalenol in the environment and food samples[J]. Journal of Hazardous Materials, 2023, 455: 131573. doi: 10.1016/j.jhazmat.2023.131573 [39] NIU C Q, XING X H, ZHANG C. A novel strategy for analyzing aptamer dominated sites and detecting AFB1 based on CRISPR–Cas12a[J]. Sensors & Diagnostics, 2023, 2(1): 155-162. [40] MA X, ZHANG Y, QIAO X J, et al. Target-Induced AIE Effect Coupled with CRISPR/Cas12a System Dual-Signal Biosensing for the Ultrasensitive Detection of Gliotoxin[J]. Analytical Chemistry, 2023, 95(31): 11723-11731. doi: 10.1021/acs.analchem.3c01760 [41] MAO Z F, WANG X J, CHEN R P, et al. Upconversion-mediated CRISPR-Cas12a biosensing for sensitive detection of ochratoxin A[J]. Talanta, 2022, 242: 123232. doi: 10.1016/j.talanta.2022.123232 [42] LIU X, BU S J, FENG J Q, et al. Electrochemical biosensor for detecting pathogenic bacteria based on a hybridization chain reaction and CRISPR-Cas12a[J]. Analytical and Bioanalytical Chemistry, 2022, 414(2): 1073-1080. doi: 10.1007/s00216-021-03733-6 [43] WU Y P, CHANG D R, CHANG Y Y, et al. Nucleic Acid Enzyme-Activated CRISPR-Cas12a With Circular CRISPR RNA for Biosensing[J]. Small, 2023, 19(41): e2303007. doi: 10.1002/smll.202303007 [44] SANTORO S W, JOYCE G F. A general purpose RNA-cleaving DNA enzyme[J]. Proceedings of the National Academy of Sciences of the United States of America, 1997, 94(9): 4262-4266. [45] LI J, ZHENG W, KWON A H, et al. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme[J]. Nucleic Acids Research, 2000, 28(2): 481-488. doi: 10.1093/nar/28.2.481 [46] GAO P, LIU B, PAN W, et al. A Spherical Nucleic Acid Probe Based on the Au–Se Bond[J]. Analytical Chemistry, 2020, 92(12): 8459-8463. doi: 10.1021/acs.analchem.0c01204 [47] CUTLER J I, AUYEUNG E, MIRKIN C A. Spherical Nucleic Acids[J]. Journal of the American Chemical Society, 2012, 134(3): 1376-1391. doi: 10.1021/ja209351u [48] CHEN Y, CHEN L, OU Y, et al. DNAzyme-based biosensor for Cu2+ ion by combining hybridization chain reaction with fluorescence resonance energy transfer technique[J]. Talanta, 2016, 155: 245-249. doi: 10.1016/j.talanta.2016.04.057 [49] SARAN R, LIU J W. A Silver DNAzyme[J]. Analytical Chemistry, 2016, 88(7): 4014-4020. doi: 10.1021/acs.analchem.6b00327 [50] ZHOU W H, VAZIN M, YU T M, et al. In vitro selection of chromium-dependent DNAzymes for sensing chromium(III) and chromium(VI)[J]. Chemistry, 2016, 22(28): 9835-9840. doi: 10.1002/chem.201601426 [51] Wu Y T, Torabi S F, Lake R J, et al. Simultaneous Fe2+/Fe3+ imaging shows Fe3+ over Fe2+ enrichment in Alzheimer's disease mouse brain[J]. Science Advances, 2023, 9(16): eade7622. doi: 10.1126/sciadv.ade7622 [52] 郑星, 马明, 崔力, 等. 应用于环境分析的切割RNA的脱氧核酶研究进展[J]. 化学通报, 2022, 85(7): 770-780. ZHENG X, MA M, CUI L, et al. Advances in RNA-Cleaving DNAzymes for environmental analysis[J]. Chemistry, 2022, 85(7): 770-780 (in Chinese).
[53] RAJENDRAN M, ELLINGTON A D. Selection of fluorescent aptamer beacons that light up in the presence of zinc[J]. Analytical and Bioanalytical Chemistry, 2008, 390(4): 1067-1075. doi: 10.1007/s00216-007-1735-8 [54] WANG H Y, CHENG H, WANG J N, et al. Selection and characterization of DNA aptamers for the development of light-up biosensor to detect Cd(II)[J]. Talanta, 2016, 154: 498-503. doi: 10.1016/j.talanta.2016.04.005 [55] WU Y G, ZHAN S S, WANG L M, et al. Selection of a DNA aptamer for cadmium detection based on cationic polymer mediated aggregation of gold nanoparticles[J]. Analyst, 2014, 139(6): 1550-1561. doi: 10.1039/C3AN02117C [56] CHEN Y, LI H H, GAO T, et al. Selection of DNA aptamers for the development of light-up biosensor to detect Pb(II)[J]. Sensors and Actuators B: Chemical, 2018, 254: 214-221. doi: 10.1016/j.snb.2017.07.068 [57] WRZESINSKI J, CIESIOLKA J. Characterization of structure and metal ions specificity of Co2+-binding RNA aptamers[J]. Biochemistry, 2005, 44(16): 6257-6268. doi: 10.1021/bi047397u [58] QU H, CSORDAS A T, WANG J P, et al. Rapid and Label-Free Strategy to Isolate Aptamers for Metal Ions[J]. ACS Nano, 2016, 10(8): 7558-7565. doi: 10.1021/acsnano.6b02558 [59] 王斌, 邓述波, 黄俊, 等. 我国新兴污染物环境风险评价与控制研究进展[J]. 环境化学, 2013, 32(7): 1129-1136. doi: 10.7524/j.issn.0254-6108.2013.07.003 WANG B, DENG S B, HUANG J, et al. Environmental risk assessment and control of emerging contaminants in china[J]. Environmental Chemistry, 2013, 32(7): 1129-1136 (in Chinese). doi: 10.7524/j.issn.0254-6108.2013.07.003
[60] ZHUO Z J, YU Y Y, WANG M L, et al. Recent Advances in SELEX technology and aptamer applications in biomedicine[J]. International Journal of Molecular Sciences, 2017, 18(10): 2142. doi: 10.3390/ijms18102142 [61] 陈慧甜, 孙清, 时国庆, 等. 核酸适配体在环境分析中的应用[J]. 环境化学, 2015, 34(1): 89-96. doi: 10.7524/j.issn.0254-6108.2015.01.2014101401 CHEN H T, SUN Q, SHI G Q, et al. Application of aptamers to environmental analysis[J]. Environmental Chemistry, 2015, 34(1): 89-96 (in Chinese). doi: 10.7524/j.issn.0254-6108.2015.01.2014101401
[62] 孟雪洁, 张瑜, 刘京华, 等. 纳米金-适配体电化学传感器用于环境水样中双酚A检测[J]. 环境化学, 2023, 42(2): 379-387. doi: 10.7524/j.issn.0254-6108.2021102403 MENG X J, ZHANG Y, LIU J H, et al. Gold nanoparticles-aptamer electrochemical sensor for detection of bisphenol A in environmental waters[J]. Environmental Chemistry, 2023, 42(2): 379-387 (in Chinese). doi: 10.7524/j.issn.0254-6108.2021102403
[63] LIU H M, LU A X, FU H L, et al. Affinity capture of aflatoxin B(1) and B(2) by aptamer-functionalized magnetic agarose microspheres prior to their determination by HPLC[J]. Mikrochimica Acta, 2018, 185(7): 326. doi: 10.1007/s00604-018-2849-8 [64] CAI S D, YAN J H, XIONG H J, et al. Investigations on the interface of nucleic acid aptamers and binding targets[J]. Analyst, 2018, 143(22): 5317-5338. doi: 10.1039/C8AN01467A [65] LEI Z, LEI P, GUO J F, et al. Recent advances in nanomaterials-based optical and electrochemical aptasensors for detection of cyanotoxins[J]. Talanta, 2022, 248: 123607. doi: 10.1016/j.talanta.2022.123607 [66] MONDAL B, RAMLAL S, LAVU P S R, et al. A combinatorial systematic evolution of ligands by exponential enrichment method for selection of aptamer against protein targets[J]. Applied Microbiology and Biotechnology, 2015, 99(22): 9791-9803. doi: 10.1007/s00253-015-6858-9 [67] PARK J Y, YANG K A, CHOI Y J, et al. Novel ssDNA aptamer-based fluorescence sensor for perfluorooctanoic acid detection in water[J]. Environment International, 2022, 158: 107000. doi: 10.1016/j.envint.2021.107000 [68] CHEN J H, SHI G, YAN C. Visual test paper for on-site polychlorinated biphenyls detection and its logic gate applications[J]. Analytical Chemistry, 2021, 93(46): 15438-15444. doi: 10.1021/acs.analchem.1c03309 [69] CHEN Y Q, WANG Z M, LIU S Y, et al. A highly sensitive and group-targeting aptasensor for total phthalate determination in the environment[J]. Journal of Hazardous Materials, 2021, 412: 125174. doi: 10.1016/j.jhazmat.2021.125174 [70] 高羽菲, 甄建辉, 赵杰, 等. 比色法适配体传感器在抗生素检测中的研究进展 [J]. 分析实验室, 2023, 1-18. GAO Y F, ZHEN J H, ZHAO J, et al. Advances in antibiotics detection based on colorimetric aptasensors[J]. Chinese Journal of Analysis Laboratory, 2023, 1-18 (in Chinese).
[71] XU G H, WANG C, YU H, et al. Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer[J]. Nucleic Acids Research, 2023, 51(14): 7666-7674. doi: 10.1093/nar/gkad541 [72] MCCONNELL E M, NGUYEN J, LI Y F. Aptamer-based biosensors for environmental monitoring[J]. Frontiers in Chemistry, 2020, 8: 434. doi: 10.3389/fchem.2020.00434 [73] CHANG T J, HE S S, AMINI R, et al. Functional nucleic acids under unusual conditions[J]. ChemBioChem, 2021, 22(14): 2368-2383. doi: 10.1002/cbic.202100087 [74] ACKERMAN C M, MYHRVOLD C, THAKKU S G, et al. Massively multiplexed nucleic acid detection with Cas13[J]. Nature, 2020, 582(7811): 277-282. doi: 10.1038/s41586-020-2279-8 [75] ALI M M, WOLFE M, TRAM K, et al. A DNAzyme‐Based colorimetric paper sensor for helicobacter pylori[J]. Angewandte Chemie International Edition, 2019, 58(29): 9907-9911. doi: 10.1002/anie.201901873 [76] ROTHENBROKER M, MCCONNELL E M, GU J, et al. Selection and Characterization of an RNA-Cleaving DNAzyme Activated by Legionella pneumophila[J]. Angewandte Chemie International Edition, 2021, 60(9): 4782-4788. doi: 10.1002/anie.202012444 [77] ZHOU Q B, ZHANG G X, WU Y P, et al. In Vitro selection of M2+-independent, fast-responding acidic deoxyribozymes for bacterial detection[J]. Journal of the American Chemical Society, 2023, 145(39): 21370-21377. doi: 10.1021/jacs.3c06155 [78] CHANG T J, LI G P, CHANG D R, et al. An RNA‐Cleaving DNAzyme that requires an organic solvent to function[J]. Angewandte Chemie International Edition, 2023, 62(42): e202310941. doi: 10.1002/anie.202310941 -




 下载:
下载: