-
近年来,水体咸化富营养化问题日益严重,水资源短缺问题日益突出。人工湿地作为一种水体生态修复技术,可应用于咸化富营养化水体的治理[1-3]。有研究[4-5]表明,人工湿地系统处理含盐废水的脱氮除磷效果较好。
植物是构成人工湿地的重要部分,湿地植物根系存在根际效应,根际中微生物数量和活性高于非根际环境,可以促进湿地处理系统的硝化和反硝化作用,强化湿地生物脱氮能力[6]。有研究[7]表明,根际周围的微生物通常是非根际的几十到几千倍,很多有机物高效降解菌株是由植物根际分离获得的。然而盐度胁迫会抑制微生物的生长,高锋等[8]将人工湿地应用于含盐生活污水处理,当进水盐度达到2.00%时,人工湿地基质中细菌、真菌、放线菌和硝化细菌的数量明显减少,基质脲酶、纤维素酶活性也相应下降,湿地基质微生物数量及活性受到抑制。
目前人工湿地处理含盐富营养化水的微生态学机理研究较少,尤其对不同盐度水平下的湿地植物根际与非根际的微生物菌群变化尚未探究。因此,本研究构建了模拟千屈菜垂直流人工湿地系统,处理盐度为0.05%、0.50%、1.00%的富营养化水,并通过高通量测序分析了不同盐度下千屈菜根际与非根际微生物菌群变化,研究结果对揭示垂直流人工湿地处理含盐富营养化水微生态学机理具有参考价值。
全文HTML
-
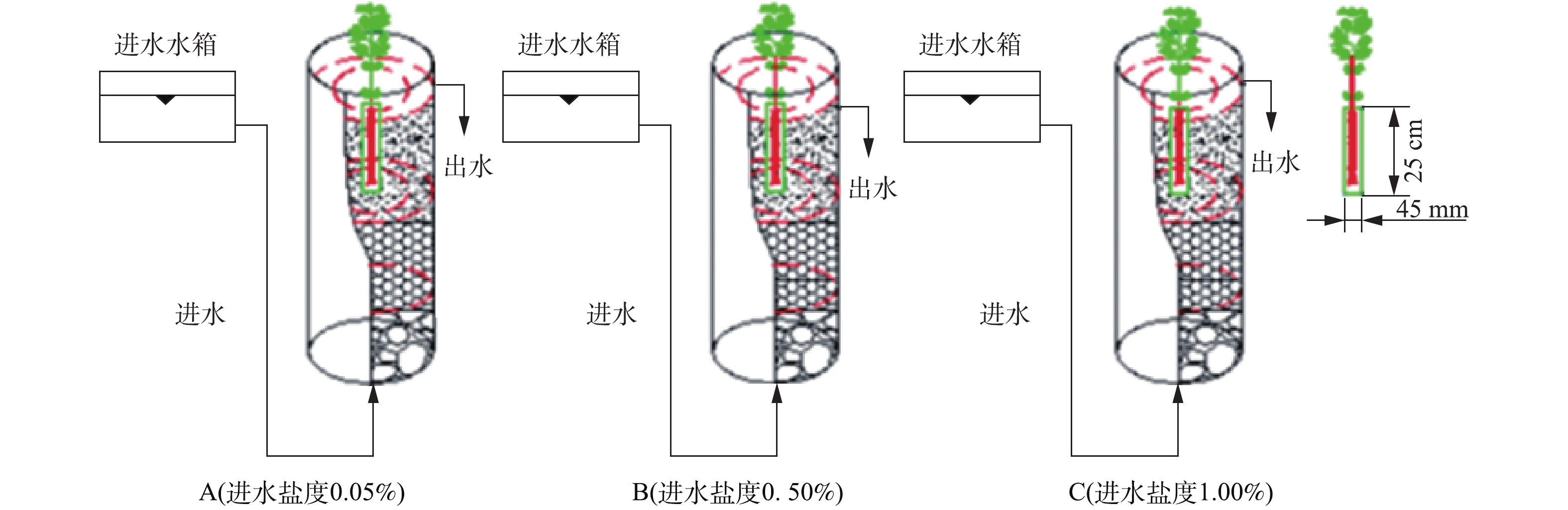
模拟垂直流人工湿地系统实验装置为3根内径150 mm、高700 mm的有机玻璃圆柱,分别记为A(进水盐度0.05%)、B(进水盐度0.50%)、C(进水盐度1.00%),示意图见图1。
柱内从下到上依次填充不同填料:最下层(100 mm)为平均粒径为30~40 mm的碎石;中层(300 mm)为平均粒径为10~20 mm的陶粒;最上层(200 mm)为土壤层。所用土壤取自天津大学北洋园校区绿化带。为区分根际(rhizosphere,RS)与非根际(non-rhizosphere,NRS)环境,在A、B、C柱中位置分别放置高250 mm、直径45 mm的根袋[9]。根袋为透水500目白色尼龙膜,根袋位于柱中央,其口部与土壤表面平齐,植物根系全部埋入土壤,构成完整的模拟垂直流湿地系统。
-
根据前期研究成果,选择耐盐阈值为1.00%的挺水植物千屈菜为受试植物[9]。植物购于江苏宿迁水生植物中心,选择长势相当的3株千屈菜分别种植于柱A、B、C的根袋中,保证千屈菜根系全部位于根袋中。
-
进水为模拟含盐富营养化水,在自来水中添加各种试剂配制而成,配水中添加葡萄糖作为微生物生长所需碳源,以硝酸钾作为氮源,以二水合磷酸二氢钠和十二水合磷酸氢二钠作为磷源,水质指标平均值见表1。模拟垂直流湿地反应器为连续上向流,采用高位水箱自底部进水,柱顶部出水。运行期间,水力负荷为0.12 m3·(m2·d)−1,实际水力停留时间(HRT)为24 h,名义HRT为141.6 h。模拟垂直流湿地运行前25 d,进水中均不加NaCl;运行的第26~60天,B、C柱进水中添加NaCl,分别调整进水盐度为0.50%与1.00%,A柱为不添加NaCl的自来水,测定其盐度为0.05%。
-
每天取进出水水样后,在2 h内测定水中的TN、
$ {\rm{NH}}_{\rm{4}}^{\rm{ + }}$ -N、硝态氮($ {\rm{NO}}_{\rm{3}}^{\rm{ - }}$ -N),参照国家环境保护局发布的标准方法[10]测定。氨氮($ {\rm{NH}}_{\rm{4}}^{\rm{ + }}$ -N):纳氏试剂分光光度比色法;亚硝酸盐氮($ {\rm{NO}}_{\rm{2}}^{\rm{ - }}$ -N):N-(1-萘基)-乙二胺分光光度法;硝酸盐氮($ {\rm{NO}}_{\rm{3}}^{\rm{ - }}$ -N):紫外分光光度法;总氮(TN):碱性过硫酸钾氧化紫外分光光度法;含盐量使用便携式电导率仪(DDBJ-350)测定;pH采用JENCO便携式酸度计测定;DO用哈希便携式溶氧仪测定。 -
在样品采集时,使用75%酒精擦洗的无菌取样器分别在柱A、B、C千屈菜根际与非根际取泥样,依据千屈菜根系深度,取样深度为距土壤层表面10~15 cm处,每组根际、非根际各取5个采样点,将这5个采样点的泥样混匀后,作为一个待测样品冻存于−20 ℃中。A、B、C柱内千屈菜根际与非根际环境共取得6个样品,分别记为NRS0.05(非根际,盐度0.05%)、RS0.05(根际,盐度0.05%)、NRS0.50(非根际,盐度0.50%)、RS0.50(根际,盐度0.50%)、NRS1.00(非根际,盐度1.00%)、RS1.00(根际,盐度1.00%)。
样品均采用土壤DNA提取试剂盒(Omega Bio-Tek,Inc.,Norcross,GA,USA),提取微生物基因组DNA。之后,以16S V4区引物(515F和806R)进行PCR扩增,对所扩增的16S rRNA中的V3~V4区域,进行小片段基因文库的构建,并基于Illumina HiSeq测序平台对该文库进行双末端测序。经过Reads拼接过滤,对所有样本的有效数据以97%的一致性进行OTUs聚类,然后对OTUs的序列进行物种注释丰度分析,揭示样品物种构成并对测序结果进行分析。使用数据统计软件SPSS 26.0对数据进行独立样本t检验,P<0.05认为差异显著。
1.1. 实验装置
1.2. 植物来源与培养
1.3. 进水配水与运行参数
1.4. 分析测试方法
1.5. 样品采集与高通量测序
-
运行结果显示,盐度增加抑制了人工湿地系统的脱氮效果。由表2可知,在不同盐浓度(0.05%、0.50%、1.00%)下,A、B、C组出水TN的平均去除率分别为64.1%、55.2%、28.1%,
$ {\rm{NH}}_{\rm{4}}^{\rm{ + }}$ -N的平均去除率分别为88.2%、71.9%、59.8%,$ {\rm{NO}}_{\rm{3}}^{\rm{ - }}$ -N的平均去除率分别为93.8%、100%和95.8%。随着盐浓度的提高,TN和$ {\rm{NH}}_{\rm{4}}^{\rm{ + }}$ -N的去除率明显降低,TN去除率低的主要原因是硝化过程受到抑制,$ {\rm{NH}}_{\rm{4}}^{\rm{ + }}$ -N无法转化为$ {\rm{NO}}_{\rm{3}}^{\rm{ - }}$ -N,这可能是因为盐浓度的提高抑制了系统中硝化菌的活性[11]。在各盐度水平下,$ {\rm{NO}}_{\rm{3}}^{\rm{ - }}$ -N的去除率均大于95%。这表明,湿地系统内的反硝化细菌在盐胁迫下仍能完成对系统内$ {\rm{NO}}_{\rm{3}}^{\rm{ - }}$ -N的转化,盐度胁迫下反硝化菌更能适应环境。郭姿璇等[12]在探究盐度对微生物活性的影响实验中同样发现反硝化菌比硝化菌的盐耐受性更强。 -
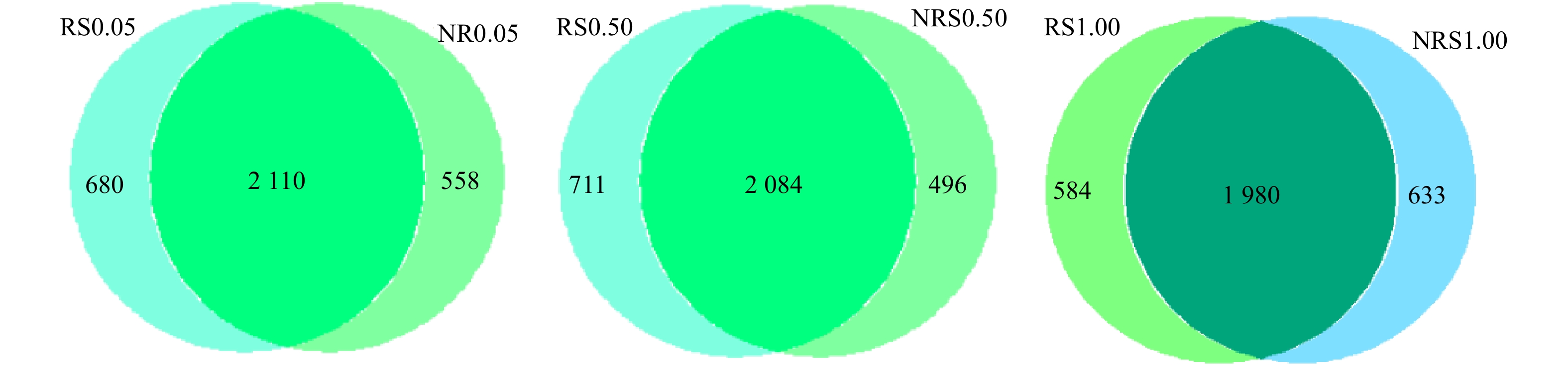
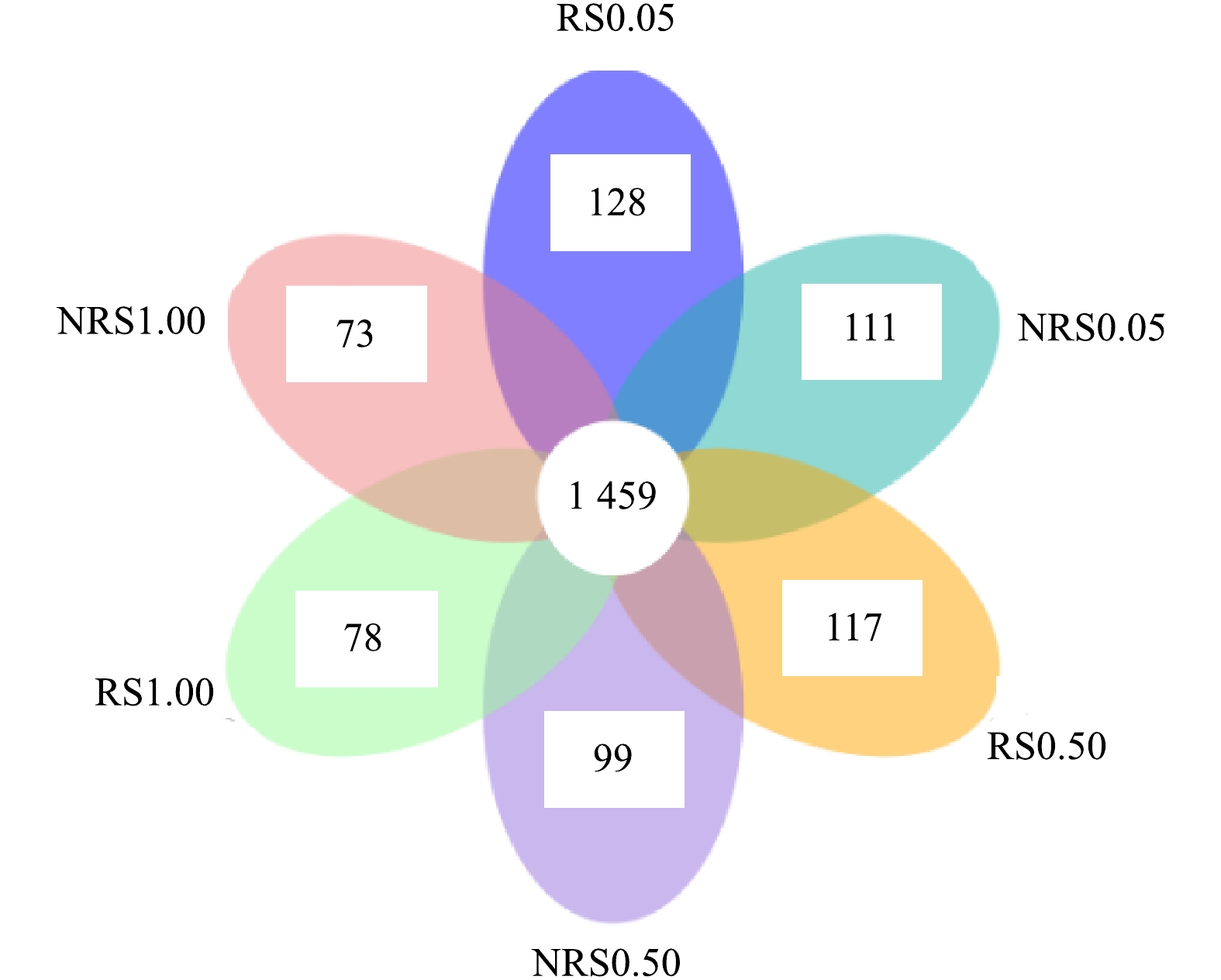
分析NRS0.05(非根际,盐度0.05%)、RS0.05(根际,盐度0.05%)、NRS0.50(非根际,盐度0.50%)、RS0.50(根际,盐度0.50%)、NRS1.00(非根际,盐度1.00%)、RS1.00(根际,盐度1.00%)6个样品之间共有、特有的OTUs,得出盐度水平分别为0.05%、0.50%与1.00%下,千屈菜根际与非根际环境共有、特有的OTUs比较结果如图2所示。6个样品中千屈菜根际与非根际环境共有、特有的OTUs比较结果如图3所示。
如图2所示,在0.05%盐浓度下,千屈菜根际与非根际组中特有的OTU数目分别为680个和558个;在0.50%盐浓度下,千屈菜根际与非根际组中特有的OTU数目分别为711个和496个,可见在0.05%和0.50%盐浓度下,千屈菜根际组OTU数目高于非根际组且差异显著(P<0.05),这说明千屈菜根系在丰富基质微生物多样性上发挥了一定的作用。
由图3可知,盐度水平为0.05%、0.50%与1.00%时,千屈菜根际环境中特有的OTU数目分别为128、117、78个,非根际环境中特有的OTU数目分别为111、99、73个。可见随着盐度的升高,根际组样品特有的OTU数目从128个降到78个,非根际组样品特有的OTU数目从111个降到73个,千屈菜根际组特有的OTU数目高于非根际组且差异显著(P<0.05),这也说明千屈菜根系丰富了土壤中微生物的多样性。蔺中等[13]采用盆栽根袋培养法,研究了根际效应在狼尾草降解土壤污染物阿特拉津中的作用,结果表明,盆栽培养28 d后,狼尾草根际效应提高了土壤细菌、真菌和放线菌的数量和活性,这说明植物根系在丰富微生物多样性方面发挥了重要作用。
-
不同盐度水平下千屈菜根际与非根际DNA样本中Shannon指数计算结果如表3所示。可以看出,盐度水平为0.05%、0.50%与1.00%时,千屈菜根际样品中Shannon指数分别为9.535、9.642、9.519,非根际组的Shannon指数分别为9.478、9.173、9.394。根际样品中的Shannon指数均大于非根际样品且差异显著(P<0.05),说明千屈菜根系在丰富微生物多样性方面发挥了一定的作用。陈悦等[14]研究了独山子区3种优势草本植物的根际与非根际土壤微生物功能多样性,结果表明,不同植物根际土壤微生物各多样性指数均大于自身非根际土,猜测可能是因为植物根系和植物残体给根际土壤微生物提供了适宜生长的场所与物质来源,进而丰富了微生物群落多样性。潘福霞等[15]在研究水平潜流型人工湿地不同植物脱氮效果及对土壤微生物的影响实验中,同样发现种植植物可以丰富根际土壤微生物群落功能多样性。
-
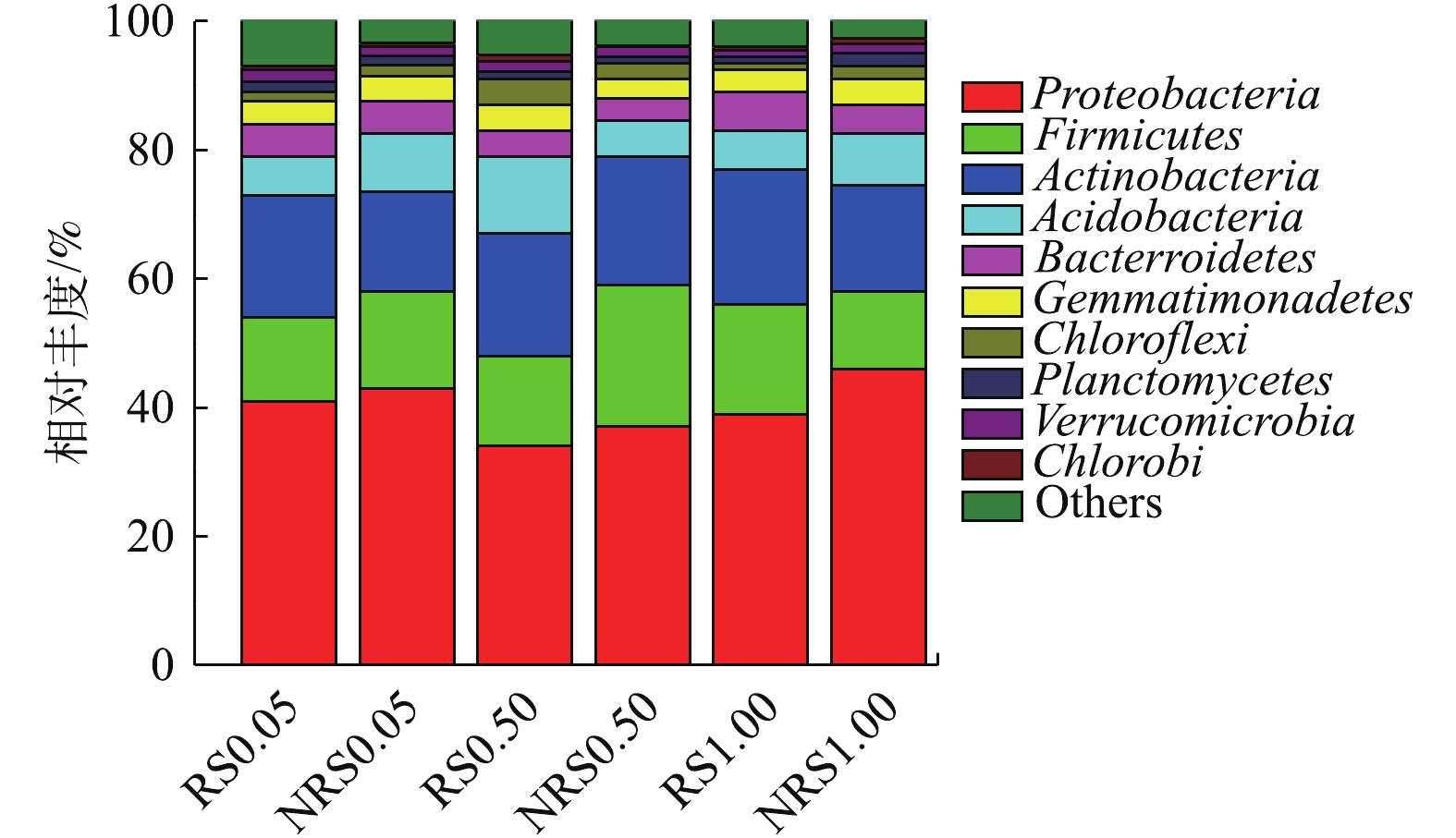
盐度水平为0.05%、0.50%与1.00%时,千屈菜根际与非根际环境中门水平上的物种丰度如图4所示。由图4可知,盐度水平为0.05%、0.50%与1.00%时,千屈菜根际与非根际样品中变形菌门(Proteobacteria)丰度最高,其次为厚壁菌门(Firmicutes)放线菌(Actinobacteria)、酸杆菌门(Acidobacteria)、拟杆菌门(Bacter roidetes)、芽单胞菌门(Bacterroidetes)。在不同盐度水平下,千屈菜根际与非根际样品在门水平上的物种差别不大,盐度与根际或非根际环境对门水平的物种相对丰度影响较小。
-
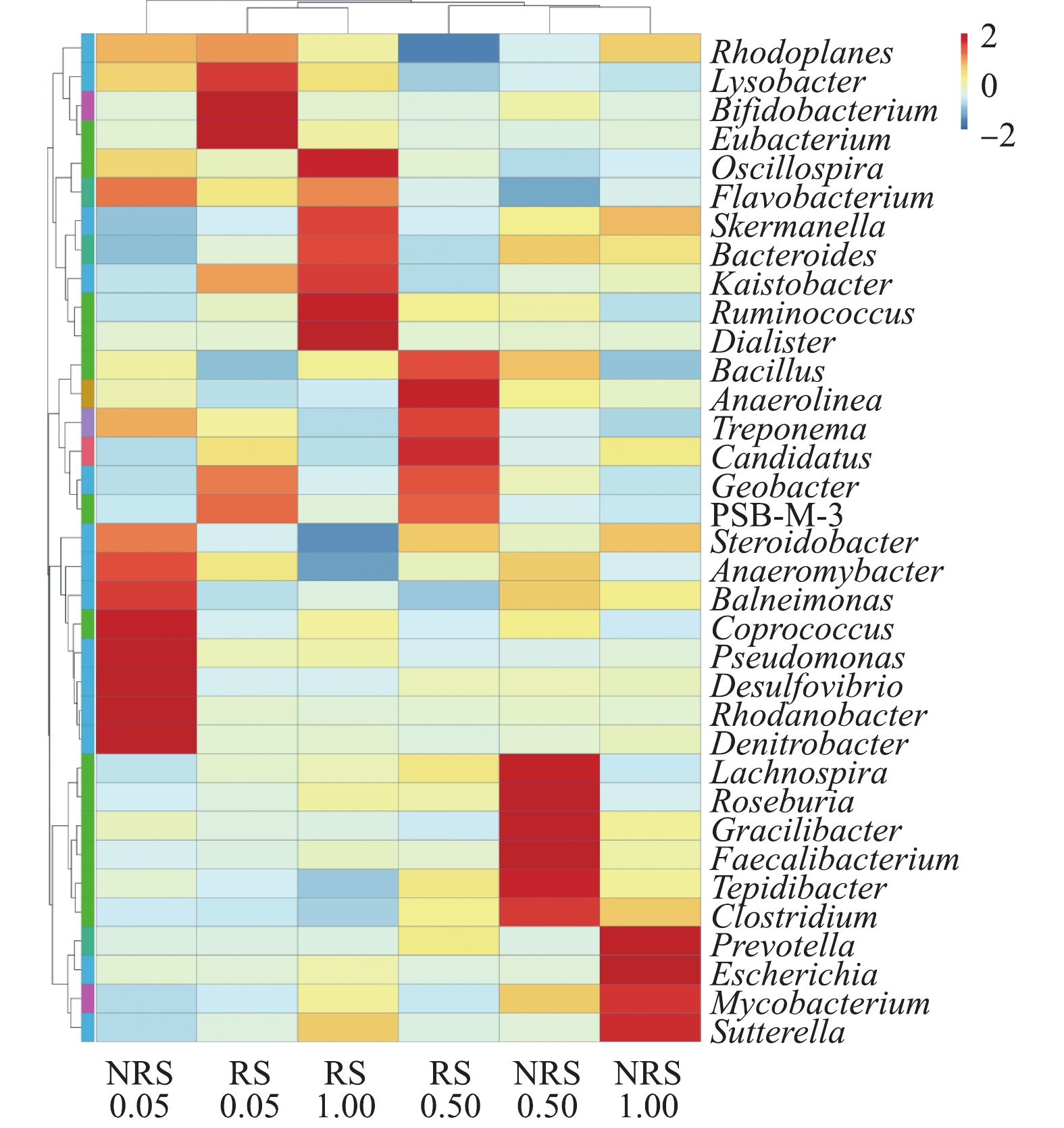
在不同盐度水平下,千屈菜根际与非根际样品中属水平上的物种丰度聚类热图如图5所示。由图5可知,盐度水平为0.05%、0.50%与1.00%时,千屈菜根际与非根际样品中的优势菌群各不相同,即使是相同盐度水平下,千屈菜根际与非根际组的菌群组成也有很大差异。
前期实验结果表明,当盐度自0.05%上升到1.00%时,
$ {\rm{NO}}_{\rm{3}}^{\rm{ - }}$ -N的去除率均大于95%。高通量分析发现,不同盐度条件下均存在反硝化功能菌,0.05%盐浓度下千屈菜非根际优势菌群中的反硝化菌种类最多。假单胞菌属Pseudomonas是典型的好氧反硝化细菌[16];红杆菌Rhodanobacter是一种兼性厌氧的革兰氏阴性杆状细菌,具有完全反硝化的能力[17];脱氮杆菌Denitrobacter是反硝化过程中的典型菌株,是一种厌氧异养反硝化菌[18]。0.05%盐浓度下千屈菜根际环境中起反硝化作用的细菌主要是真杆菌属Eubacterium,是一种严格厌氧的异养菌,可以促进反硝化,加强反硝化菌的功能[19]。在0.50%盐浓度下,千屈菜非根际中的梭菌属Clostridium是芽孢杆菌科的一属,可将硝酸盐异化还原为氨[20]。在0.50%盐浓度下,千屈菜根际中的优势菌群主要包括2种:芽孢杆菌Bacillus是一种好氧或兼性厌氧的革兰氏阳性菌,普遍存在于土壤、河道底泥中,具有丰富的酶系统和较强的代谢能力,并且能够高效脱氮,已证实其具有好氧反硝化的特性[21];地杆菌属Geobacter为专性厌氧的革兰氏阴性细菌,其可以与反硝化菌共生并促进反硝化作用[22]。
在1.00%盐浓度下,千屈菜非根际中的优势菌群种类明显减少,只检测到4种优势菌属。其中具有反硝化能力的是分支杆菌属Mycobacterium,分支杆菌属中的某些菌种有反硝化作用并有一定的耐盐性,如耻垢分枝杆菌Mycobacterium smegmatis[23]。在1.00%盐浓度下,千屈菜根际中的优势菌群种类相比非根际有明显提高且反硝化菌属种类增多,黄杆菌属Flavobacterium是一种好氧反硝化菌[24],具有一定的高盐耐受性[25-26];拟杆菌属Bacteroides为专性厌氧菌,具有反硝化作用[27]。
除了反硝化作用,前期实验结果表明:在盐胁迫下,硝化作用受到抑制,在0.50%盐度下,氨氮的平均去除率加盐后下降9%;在1.00%盐度下,氨氮去除率降低至59.8%。通过优势菌群分析,仅在0.05%盐度下的非根际组和0.50%盐度下的根际组发现有硝化作用菌群的存在。在0.05%盐浓度下,千屈菜非根际组的假单胞菌属Pseudomonas除了具有反硝化除磷功能外,某些菌种具有硝化功能,如恶臭假单胞菌Pseudomonas putida[28]和产碱假单胞菌Pseudomonas alcaligenes[29]。在0.50%盐浓度千屈菜根际组的优势菌群中,芽孢杆菌Bacillus已被证实具有异氧硝化-好氧反硝化特性,如蜡状芽孢杆菌Bacillus cereus[30-31]。Candidatus Nitrososphaera是一种氨氧化古菌(AOA),可以将氨氮氧化成亚硝酸盐[32]。在0.50%盐浓度条件下,2种硝化功能菌的存在说明千屈菜根际环境促进了硝化菌的生长。
可见,在盐度水平为0.05%、0.50%与1.00%时,千屈菜根际与非根际环境中的优势菌群皆不相同,这说明盐浓度和根际环境对微生物的群落结构产生了较大影响。仅在0.05%盐度的非根际组和0.50%盐度的根际组检测到硝化菌群的存在,可见,盐度胁迫抑制了硝化菌的生长,同时,硝化过程受到抑制,导致
$ {\rm{NH}}_{\rm{4}}^{\rm{ + }}$ -N的去除率明显降低。相比硝化菌,反硝化菌更耐盐冲击,在0.05%、0.50%与1.00%盐度水平下,均检测到反硝化菌;在1.0%盐度水平下,检测出耐盐反硝化菌黄杆菌属Flavobacterium。同时,在盐胁迫下,反硝化细菌仍能完成对系统内$ {\rm{NO}}_{\rm{3}}^{\rm{ - }}$ -N的转化,$ {\rm{NO}}_{\rm{3}}^{\rm{ - }}$ -N的去除率保持在95%以上。
2.1. 模拟湿地运行效果
2.2. 基于OTU聚类的千屈菜根际与非根际样品中Venn图和花瓣图分析
2.3. 千屈菜根际与非根际DNA样品复杂度分析
2.4. 千屈菜根际与非根际样品中门水平上细菌的物种丰度
2.5. 千屈菜根际与非根际样品中属水平上的物种丰度
-
1)模拟人工湿地系统对于含盐富营养水体的处理结果显示,盐度的增加抑制了人工湿地系统脱氮效果,尤其是TN和
$ {\rm{NH}}_{\rm{4}}^{\rm{ + }}$ -N的去除均受到了明显的抑制;然而在各盐度水平下,$ {\rm{NO}}_{\rm{3}}^{\rm{ - }}$ -N的去除率均大于95%,盐度胁迫下反硝化菌更能适应环境。2)由不同盐胁迫下OTU聚类情况分析可知,根际效应千屈菜根系丰富了土壤中微生物的多样性,根际环境中的微生物多样性明显高于非根际环境。
3)盐度的增加抑制了硝化菌的生长,仅在0.05%盐度的非根际组和0.50%盐度的根际组检测到有硝化作用菌群的存在。与硝化菌相比,反硝化菌更耐盐冲击,在0.05%、0.50%与1.00%盐度水平下,均检测到反硝化菌;在1.00%盐度水平下,检测出耐盐反硝化菌黄杆菌属Flavobacterium。





 下载:
下载: