-
石油是由烷烃、芳香烃以及少量非烃类物质(沥青和树脂等)组成的复杂混合物[1]。在石油的开采、运输、储存以及使用过程中不可避免的存在泄漏和原油落地等问题,对土壤生态环境造成不利影响[2-4]。土壤中蕴含的大量微生物通过产生对石油烃具有降解活性的酶以代谢石油烃,将其矿化为二氧化碳和水[5],对于石油污染环境的修复发挥着重要作用。石油烃降解功能基因是指微生物产生的对于石油烃具有特定降解作用的酶编码基因[6],通过对这些功能基因进行测定,可用于分析污染环境中石油烃降解菌的生物多样性,从而判定石油烃污染环境的生物修复潜势[7]。
在好氧条件下,包括芳香烃在内的许多复杂碳氢化合物较易被生物降解。然而,在非表层的石油污染土壤中,厌氧代谢是石油烃生物降解中的主要方式[8, 9]。在厌氧条件下,烷烃降解途径中琥珀酸合酶基因(masD)催化的富马酸加成反应和琥珀酸烷基代谢物的生成是烷烃降解的关键步骤[10]。而多数芳香族化合物(如甲苯、苯酚、甲酚等)在脱芳构化和裂解环之前,在微生物作用下生成中间体苯甲酰辅酶A,其中6-十六氧环-1-烯-1-羰基CoA水解酶基因(bamA)开环反应是苯甲酰辅酶A厌氧降解途径中的关键步骤[11]。因此,检测石油烃厌氧降解功能基因masD和bamA是研究土壤石油烃厌氧降解菌赋存情况、评价石油烃厌氧生物降解潜势的重要依据。
实时荧光定量聚合酶链式反应(Real-time Quantitative PCR, qPCR)是目前广为采用的基因定量检测技术[12]。在PCR体系中加入荧光探针[13]或荧光染料[14],随着扩增产物不断积累,相应的荧光信号也不断增强,通过监测荧光信号强度变化,结合标准曲线就可以定量分析未知样品中目的基因拷贝数[15-17]。在利用实时荧光定量PCR技术检测功能基因时,设计合适的扩增引物和确定退火温度是保证方法准确度和灵敏度的关键。
宋本如[18]建立了烷烃和芳烃厌氧降解关键基因masD和bamA的实时荧光定量PCR检测方法,方法可较好的用于两种基因的定量检测。但其扩增效率较低,仅为53.0%和63.6%。Aitken等[10]为研究不同条件下烷烃的活化机制,建立了扩增琥珀酸合酶基因(assA)的实时荧光定量PCR检测方法。Martijn等[19]建立了6-十六氧环-1-烯-1-羰基CoA水解酶基因(bamA)的实时荧光定量PCR检测方法,这些方法普遍存在扩增效率较低的问题。
本文通过设计引物及确定合适的退火温度和扩增程序,优化了烷烃和芳烃厌氧降解关键基因masD和bamA的定量检测方法。并利用建立的方法对陕北油井场区土壤中的masD和bamA基因进行了定量检测。研究对于评估污染土壤中石油烃的生物厌氧降解潜势具有重要的理论意义。
-
iQ5实时荧光定量PCR仪(美国Bio-Rad 公司);ND-2000微量分光光度计(美国Thermo Fisher Scientific);Mastercycler ep梯度PCR(德国Eppendorf);DYY-6D电泳仪(北京六一仪器厂);GeiDoc XR+凝胶成像仪(美国Bio-Rad公司);HR/T16MM小型离心机(湖南赫西仪器装备有限公司)。
土壤基因组DNA提取试剂盒(天根生化科技(北京)有限公司,中国);柱式质粒DNA小量抽提试剂盒(生工生物工程(上海)股份有限公司,中国);柱式DNA胶回收试剂盒(生工生物工程(上海)股份有限公司,中国);T载体PCR产物快速连接试剂盒(生工生物工程(上海)股份有限公司,中国);2×Taq PCR Master mix(KT201)(天根生化科技(北京)有限公司,中国);FastStart Essential DNA Green Master(Roche,瑞士);DH5α感受态细胞(生工生物工程(上海)股份有限公司,中国)。
-
通过查阅文献,获得可扩增(1-甲基烷基)琥珀酸合酶基因(masD)和6-十六氧环-1-烯-1-羰基CoA水解酶基因(bamA)的引物序列。引物序列、扩增产物大小、退火温度见表1。
-
按照土壤基因组DNA提取试剂盒说明程序提取采自陕北志丹县油井场区的土壤中总DNA,利用表1的masD和bamA引物扩增提取的DNA,PCR扩增体系(25 μL)为:2×Taq PCR Mastermix 12.5 μL,10.0 μmol·L−1的上下游引物各0.5 μL,DNA模板3.0 μL,ddH2O 8.5 μL。升温程序为:94 ℃预变性5 min,94 ℃变性30 s,退火温度为表1中给出的50 ℃和58 ℃,保持30 s,72 ℃延伸30 s,设定35个循环;72 ℃延伸10 min。利用1.5%琼脂糖凝胶电泳检测扩增产物的特异性。利用胶回收试剂盒回收特异性亮带,与T载体连接并克隆于DH5α感受态细胞。经过蓝白斑筛选后挑选白色菌落,置于20 mL LB培养基中[21](10 g蛋白胨、10 g NaCl和5 g酵母浸粉加入超纯水溶解,定容至1 L),30 ℃培养12—16 h,以获得的菌液为模板,通过M13通用引物(M13F:GTAAAACGACGGCCAGT;M13R:CAGGAAACAGCTATGACC)进行PCR扩增,利用1.5%琼脂糖凝胶电泳鉴定其是否为阳性克隆。最后提取阳性克隆的重组质粒送由生工生物工程(上海)股份有限公司进行测序,获得特异性亮带的DNA序列并以此为靶序列进行实时荧光定量PCR引物的设计。
-
利用Allele ID6软件根据实时荧光定量PCR引物设计原则,设计扩增masD和bamA基因的引物。具体设计原则包括:扩增产物长度在80—150 bp,最大不超过300 bp;上、下游引物Tm值相差较小,不超过2 ℃;避免模板二级结构,上、下游引物长度为18—24 bp;引物G+C含量在45%—55%之间;引物中碱基均匀并随机分布。遵循以上原则设计出实时荧光定量PCR引物masD-f2/masD-r2和bamA-f2/bamA-r2,见表2。
-
实时荧光定量PCR(20 μL)反应体系:10.0 μL的FastStart Essential DNA Green Master,上、下游引物各0.4 μL,4.2 μL RNase-Free Water,5.0 μL DNA模板,同时设置一组只加RNase-Free Water的阴性对照。
为了确定最适引物浓度和退火温度,分别向各个体系中加入0.2、0.4、0.8 μL的实时荧光定量PCR引物,使体系内引物终浓度分别为0.1、0.2、0.4 μmol·L−1。引物的退火温度分别设置为52、57、61 ℃。实时荧光定量PCR的反应程序:95 ℃预变性10 min,95 ℃变性10 s,退火温度保持15 s,72 ℃延伸10 s,共设40个循环。熔解曲线程序:以0.5 ℃·s−1由55 ℃升至95 ℃,每个温度下保持5 s。
-
用微量分光光度计测定质粒DNA的浓度(ng·µL−1),根据下式换算为copy·µL−1。将已知浓度的质粒进行8次10倍梯度稀释[22],即18 µL稀释液加入2 µL质粒稀释后,进行连续8次的梯度稀释。本研究中masD选取108—104 copy·µL−1质粒DNA共5个梯度进行标准曲线的绘制;bamA选取1010—105 copy·µL−1质粒DNA共6个梯度进行标准曲线的绘制。公式:每µL初始重组质粒中的基因绝对拷贝数(CN):
其中,X为阳性质粒浓度(ng·µL−1);T载体长为2773 bp;b为masD和bamA片段,分别长389 bp和354 bp;每对碱基的平均分子量是650 Da;阿弗加德罗常数为6.02×1023 mol−1。
利用循环阈值(Ct)与起始模板拷贝数的对数存在反比例线性关系建立标准曲线[23],作为土壤样品中masD和bamA降解基因定量的依据。
-
根据质粒DNA测序结果,利用Allele ID6软件自行设计了实时荧光定量PCR引物并优化退火温度,设计的荧光定量PCR引物序列、扩增产物大小、退火温度见表2。
-
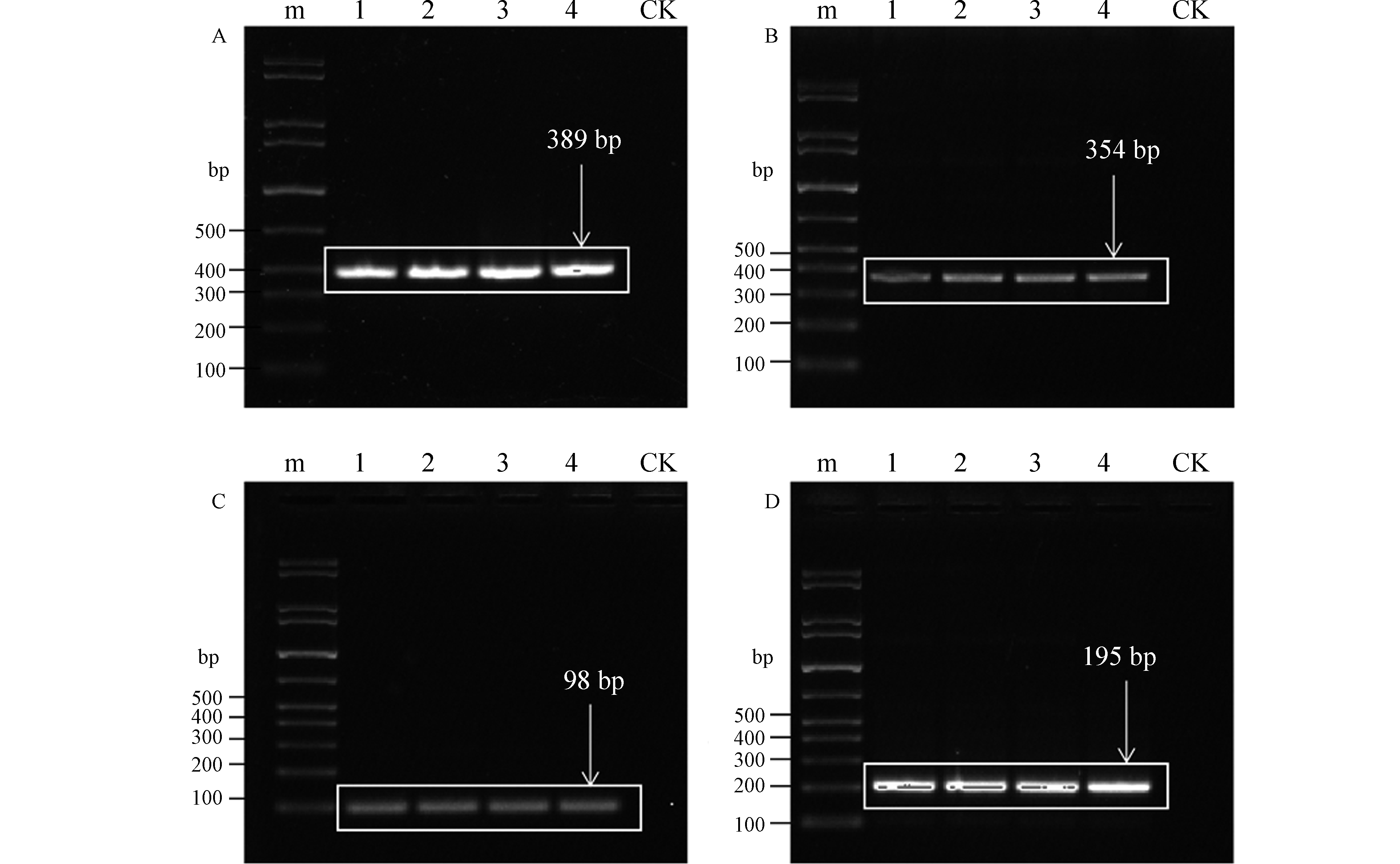
利用文献中的的masD和bamA降解基因引物(表1)对土壤总DNA进行扩增,扩增产物分别为389 bp(masD)和354 bp(bamA),产物片段较大(图1A、B)。而实时荧光定量PCR要求扩增产物不超过300 bp,本文根据质粒DNA的测序结果重新设计合成的masD和bamA降解基因的荧光定量PCR引物(表2),对masD和bamA降解基因扩增,所得产物大小为98 bp(masD)和195 bp(bamA)(图1C、D),且只有一条亮带,说明产物特异性良好,因此本文设计的引物和优化后的的反应条件与文献相比,更适合用于masD和bamA降解基因的定量检测。
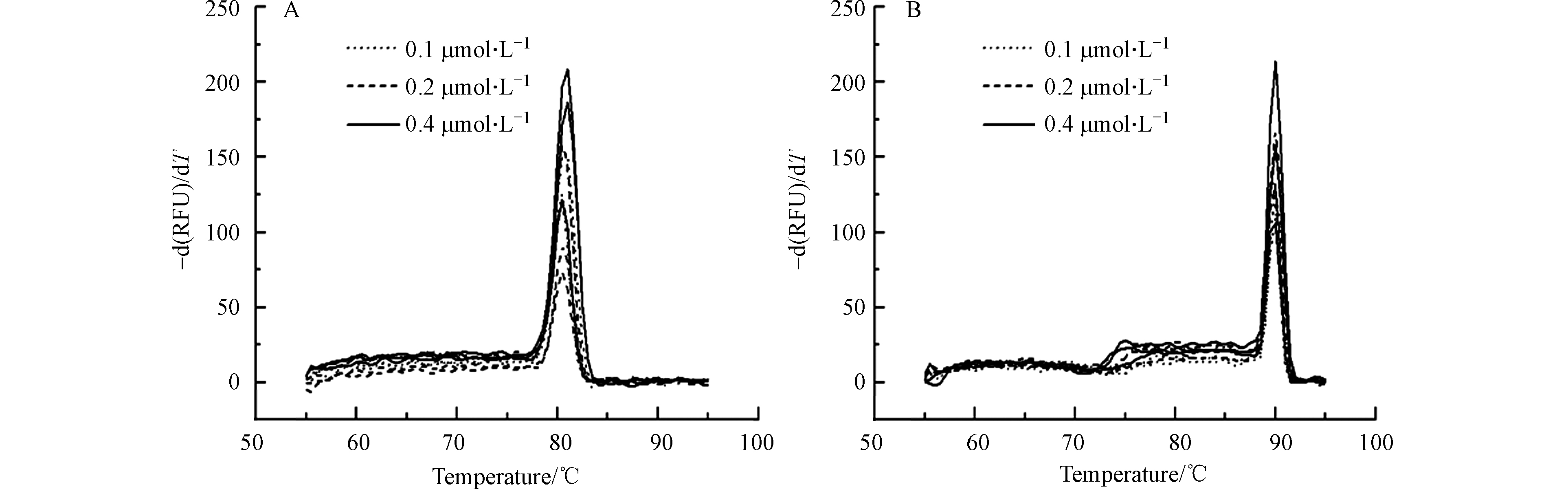
在不同引物浓度下对石油烃降解基因masD和bamA进行扩增,所得实时荧光定量PCR熔解曲线如图2所示,纵坐标为荧光强度对温度的负倒数。当引物终浓度分别为0.1、0.2、0.4 μmol·L−1时,3组熔解曲线均未出现杂峰,说明设计的引物特异性好。以产生Ct值的平均值最小和Ct值标准偏差综合指数最小为依据对引物浓度进行选择。通过比较Ct值的平均值和Ct标准偏差值,当引物终浓度为0.2 μmol·L−1时,Ct值的平均值最小且Ct值标准偏差综合指数最小(表3)。最终确定引物最适浓度为0.2 μmol·L−1。
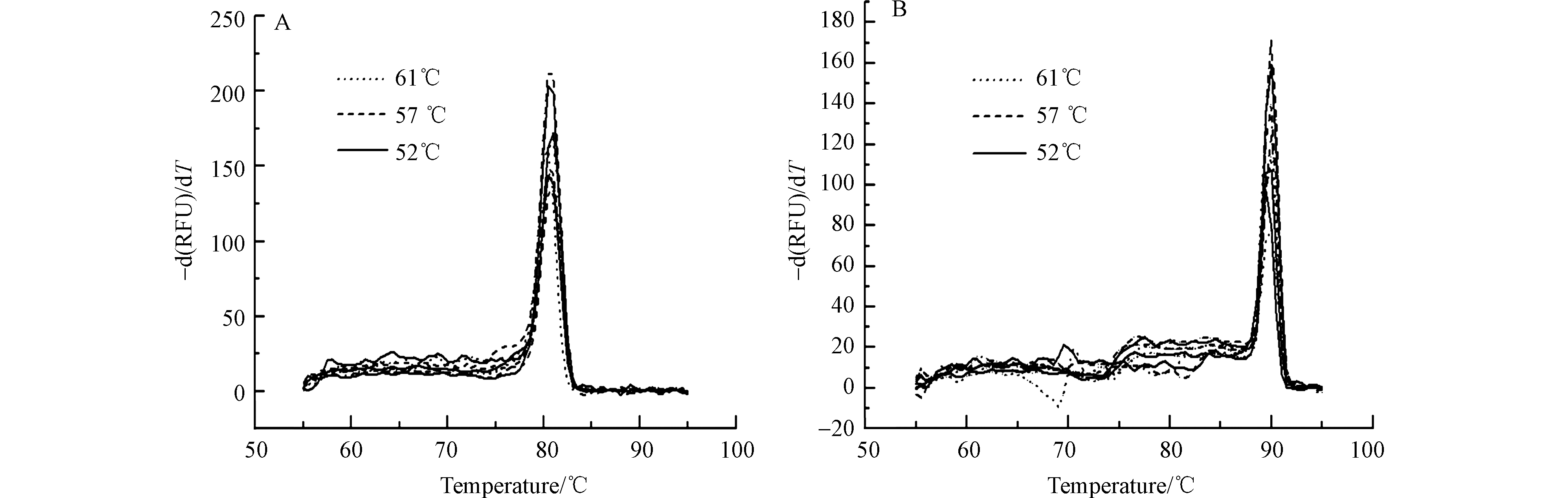
引物最适浓度确定为0.2 μmol·L−1时,石油烃厌氧关键降解基因masD和bamA在不同退火温度下的实时荧光定量PCR熔解曲线如图3所示。当退火温度分别为52、57、61 ℃时,masD和bamA降解基因的熔解曲线未出现非特异性扩增杂峰,引物显示出很高的特异性。最佳温度选择和最佳浓度选择依据的标准相同,通过比较Ct值的平均值和Ct标准偏差值,当masD基因退火温度为61 ℃、bamA基因退火温度为57 ℃时,Ct值的平均值最小且Ct值标准偏差综合指数最小。因此经优化后,最终确定体系中引物终浓度为0.2 μmol·L−1,引物masD-f/masD-r和bamA-f/bamA-r退火温度分别为61 ℃和57 ℃(表4)。
-
如图4所示,masD降解基因的标准曲线是Ct=-3.383lgC+39.022,相关系数R2=1.000,扩增效率E=97.5%;bamA降解基因的标准曲线是Ct=-4.283lgC+50.520,相关系数R2=0.993,扩增效率E=71.2%。文献报道测定masD和bamA方法中的扩增效率分别为53.0%和63.6%[18],该方法与文献报道方法相比,提高了7.6%—44.5%。本文通过设计扩增引物并优化扩增条件,使扩增效率显著提高,提高了方法的灵敏度和准确性。
-
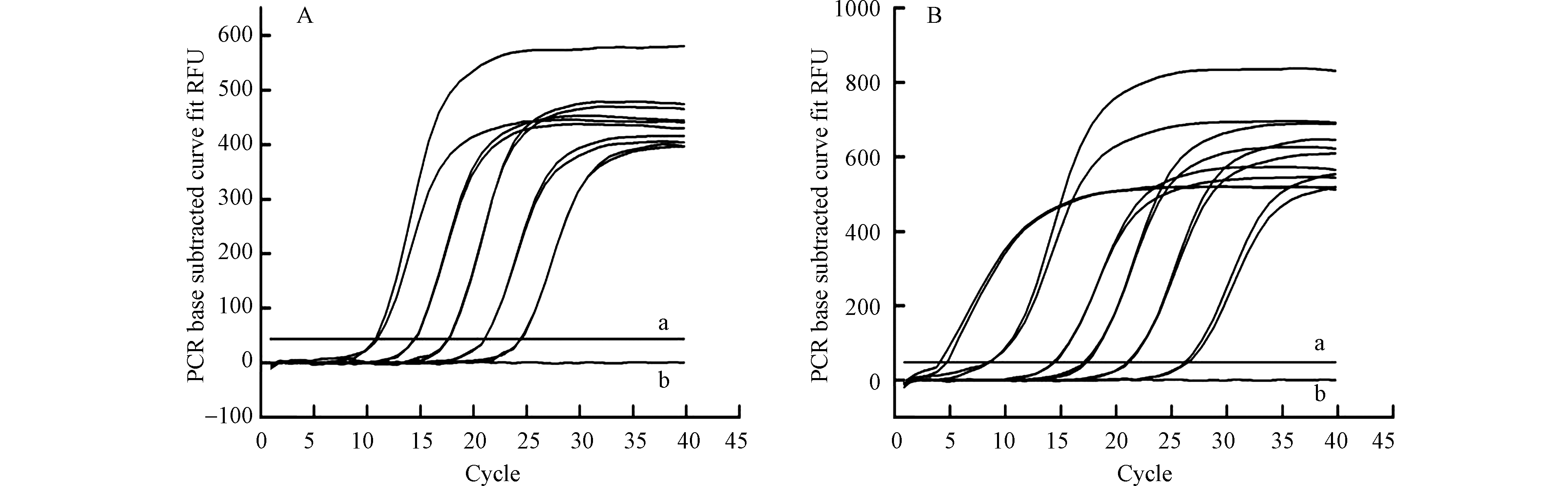
如图5所示,降解基因masD和bamA同一浓度梯度下的两条扩增曲线几乎重合,组内变异系数除bamA降解基因1010浓度梯度外均小于1%(表5),说明利用所设计的引物对两种基因进行实时荧光定量PCR检测时,方法的重复性好,可保证不同样品间检测结果的可靠性和稳定性[24]。
-
利用所建立的实时定量荧光PCR方法定量检测了陕北5个地区石油污染土壤中的masD和bamA降解基因的拷贝数,结果如表6所示,5个地区石油污染土壤的masD和bamA降解基因拷贝数分别在2.69×102—7.52×103 copy·g−1 土及1.00×104—3.60×105 copy·g−1 土之间,土壤中bamA降解基因的拷贝数大于masD降解基因,两种基因拷贝数与土壤中石油烃含量无相关关系。在5个地区污染土壤中均可检测到两种控制石油烃厌氧降解的关键基因,说明在石油污染土壤中蕴含着可对石油烃进行厌氧降解代谢的微生物,有可能通过厌氧修复的方式对石油污染土壤进行修复治理。
-
本论文通过设计检测石油烃厌氧降解关键基因masD和bamA的定量扩增引物,并优化反应条件和扩增程序,建立了利用实时荧光定量PCR技术对烷烃厌氧降解基因masD和芳香烃厌氧降解基因bamA进行定量检测的方法。与文献报道的方法相比,优化后的方法具有更好的重复性、灵敏度和扩增效率。利用建立的方法对陕北石油开采区污染土壤进行定量检测的结果表明,石油污染土壤中普遍存在着控制石油烃厌氧降解的关键基因,说明在石油污染土壤中蕴含着可对石油烃厌氧代谢的微生物,在对石油污染土壤进行修复时,有望通过厌氧生物降解的方式对污染土壤进行修复治理。
石油烃厌氧降解关键基因masD和bamA的实时荧光定量PCR检测方法与应用
Real-time quantitative PCR for determination of petroleum hydrocarbon anaerobic degradation key genes masD and bamA
-
摘要: masD和bamA是控制石油烃厌氧降解的关键基因,利用实时荧光定量PCR技术检测masD和bamA基因具有简便快速和易操作等优点。但目前所用方法存在扩增效率低,方法灵敏度较差的问题。本文根据引物设计原则,利用Allele ID6软件重新设计了扩增masD和bamA的实时荧光定量PCR引物,将质粒DNA进行8次10倍梯度稀释后构建实时荧光定量PCR标准曲线。优化后的体系(20 μL)为:FastStart Essential DNA Green Master 10.0 μL,上下游引物各0.4 μL,RNase-Free Water 4.2 μL,5.0 μL DNA 模板。利用新设计的引物扩增masD和bamA基因的最适退火温度分别为61 ℃和57 ℃。优化后的检测方法扩增效率提高至97.5%和71.2%,比文献报道的方法提高了7.6%—44.5%,具有更高的重复性和灵敏度。利用设计的引物对陕北5个地区石油污染土壤中的masD和bamA基因进行定量检测结果表明,石油污染土壤中普遍存在着控制石油烃厌氧降解的关键基因,所测定的土壤中bamA降解基因的拷贝数远高于masD降解基因。Abstract: The masD and bamA are key genes controlling the anaerobic degradation of petroleum hydrocarbons. Real-time quantitative PCR (qPCR) is the common gene detection method which has the advantages of simple, fast and easy operation. In this study, according to the primer design principle, the qPCR primers for amplifying masD and bamA were designed using Allele ID6 software. The obtained primers have uniform base distribution, appropriate amplification length, minor difference of Tm values between forward and reverse primers, as well as single peak in the fusion curve of amplification products, indicating that the primers had good specificity. The plasmid DNA was diluted 8 times with 10 fold gradient to construct the qPCR standard curve. 20 μL of optimized system included 10.0 μL of Faststart essential DNA green master, 4.2 μL RNase-Free Water, 5.0 μL DNA template, 0.4 μL of upstream and downstream primers, respectively. The optimum annealing temperatures for amplification of masD and bamA genes were 61 ℃ and 57 ℃, respectively. Compared with the literatures, the amplification efficiency of this optimized method enhanced by 7.6%—44.5%, indicating the method has higher repeatability and sensitivity. This optimized method can be used to quantitatively detect the masD and bamA genes in the petroleum-contaminated soils collected from five regions in northern of Shaanxi province of China. The results indicated that the anaerobic degradation genes of masD and bamA were ubiquitous in the petroleum-contaminated soils, and the gene copy numbers of bamA was much higher than that of masD in the oil-polluted soils.
-

-
表 1 文献报道的masD和bamA降解基因引物
Table 1. Primers of masD and bamA degradation genes reported in literature
表 2 本文设计的扩增masD和bamA降解基因的荧光定量PCR引物
Table 2. Self-designed fluorescent quantitative PCR primers for masD and bamA degradation genes
引物
Primers引物序列
Sequence (5’-3’)扩增尺寸/ bp
Amplicon size退火温度/℃
Annealing temperaturemasD-f2 GCTGAAGAAAGGCTGCGTTA 98 61 masD-r2 TGATACTTACGGGCAGACCA bamA-f2 TGATATGGTGAAGGCGGTGA 195 57 bamA-r2 GCCGAAACCATGTCGTTGAA 表 3 引物浓度优化数据
Table 3. Primer concentration optimization data
引物终浓度/(µmol·L−1)
Final primer concentrationmasD bamA Ct平均值
Ct MeanCt标准偏差
Ct SDCt平均值
Ct MeanCt标准偏差
Ct SD0.1 18.47 0.104 28.95 0.799 0.2 13.79 0.069 23.09 0.044 0.4 14.93 0.357 24.45 0.103 表 4 退火温度优化数据
Table 4. Annealing temperature optimization data
退火温度/℃
Annealing temperaturemasD bamA Ct平均值
Ct MeanCt标准偏差
Ct SDCt平均值
Ct MeanCt标准偏差
Ct SD52 18.95 0.079 27.98 0.619 57 19.42 0.133 25.11 0.105 61 18.91 0.036 26.10 0.153 表 5 实时荧光定量PCR检测masD和bamA基因的组内重复性试验结果
Table 5. Intra- repeatability of masD and bamA genes detected by real-time fluorescence quantitative PCR
质粒标准品浓度/(copy·μL−1)
Plasmid standard concentrationmasD bamA 组内变异系数
Coefficient of variation within groups组内变异系数
Coefficient of variation within groupsCt`x±SD 变异系数/%
Coefficient of variationCt`x±SD 变异系数/%
Coefficient of variation1010 4.44 ±0.29 6.49% 109 8.52 ±0.02 0.19% 108 10.88 ±0.10 0.90% 14.36 ±0.06 0.45% 107 14.43 ±0.04 0.29% 17.21 ±0.11 0.64% 106 17.59 ±0.06 0.35% 21.07 ±0.05 0.26% 105 21.06 ±0.01 0.06% 26.32 ±0.15 0.56% 104 24.48 ±0.05 0.21% 表 6 陕北油田区污染土壤中masD和bamA降解基因定量结果
Table 6. Quantitative results of masD and bamA degradation genes in five polluted soils in the northern of Shaanxi province
地区
Regions石油烃浓度/(mg·kg−1 土)
Petroleum hydrocarbon concentrationsmasD 拷贝数/(copy·g−1土)
masD copy numbersbamA 拷贝数/(copy·g−1土)
bamA copy numbers安塞Ansai 50200 7.52×103 1.00×104 志丹Zhidan 26633 4.40×103 3.05×104 延安Yanan 6661 4.65×102 5.05×104 绥德Suide 5533 2.69×102 3.60×105 定边Dingbian 146500 4.43×102 1.43×105 -
[1] VARJANI S J. Microbial degradation of petroleum hydrocarbons [J]. Bioresource Technology, 2017, 223: 277-286. doi: 10.1016/j.biortech.2016.10.037 [2] 乔俊, 陈威, 张承东. 添加不同营养助剂对石油污染土壤生物修复的影响 [J]. 环境化学, 2010, 29(1): 6-11. QIAO J, CHEN W, ZHANG C D. Bioremediation of petroleum contaminated soil by various nutrient amendments [J]. Environmental Chemistry, 2010, 29(1): 6-11(in Chinese).
[3] 杨茜, 吴蔓莉, 聂麦茜, 等. 石油污染土壤的生物修复技术及微生物生态效应 [J]. 环境科学, 2015, 36(5): 1856-1863. YANG Q, WU M L, NIE M Q, et al. Effects and biological response on bioremediation of petroleum contaminated soil [J]. Environmental Science, 2015, 36(5): 1856-1863(in Chinese).
[4] WU M L, LI W, DICK W A, et al. Bioremediation of hydrocarbon degradation in a petroleum-contaminated soil and microbial population and activity determination [J]. Chemosphere, 2017, 169: 124-130. doi: 10.1016/j.chemosphere.2016.11.059 [5] 李洲. 微生物对石油烃的降解机理研究 [J]. 云南化工, 2018, 45(9): 179-180. doi: 10.3969/j.issn.1004-275X.2018.09.081 LI Z. Study on the mechanism of microbial degradation of petroleum hydrocarbons [J]. Yunnan Chemical Technology, 2018, 45(9): 179-180(in Chinese). doi: 10.3969/j.issn.1004-275X.2018.09.081
[6] 吴慧君, 宋权威, 郑瑾, 等. 微生物降解石油烃的功能基因研究进展 [J]. 微生物学通报, 2020, 47(10): 3355-3368. WU H J, SONG Q W, ZHENG J, et al. Function genes in microorganisms capable of degrading petroleum hydrocarbon [J]. Microbiology China, 2020, 47(10): 3355-3368(in Chinese).
[7] SHAHI A, INCE B, AYDIN S, et al. Assessment of the horizontal transfer of functional genes as a suitable approach for evaluation of the bioremediation potential of petroleum-contaminated sites: A mini-review [J]. Applied Microbiology and Biotechnology, 2017, 101(11): 4341-4348. doi: 10.1007/s00253-017-8306-5 [8] AITKEN C M, JONES D M, LARTER S R. Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs [J]. Nature, 2004, 431(7006): 291-294. doi: 10.1038/nature02922 [9] WARTELL B, BOUFADEL M, RODRIGUEZ-FREIRE L. An effort to understand and improve the anaerobic biodegradation of petroleum hydrocarbons: A literature review [J]. International Biodeterioration & Biodegradation, 2021, 157: 105156. [10] AITKEN C M, JONES D M, MAGUIRE M J, et al. Evidence that crude oil alkane activation proceeds by different mechanisms under sulfate-reducing and methanogenic conditions [J]. Geochimica et Cosmochimica Acta, 2013, 109: 162-174. doi: 10.1016/j.gca.2013.01.031 [11] LI Y N, LI J, WANG D, et al. Denitrifying microbial community structure and BamA gene diversity of phenol degraders in soil contaminated from the coking process [J]. Applied Biochemistry and Biotechnology, 2020, 190(3): 966-981. doi: 10.1007/s12010-019-03144-5 [12] NAVARRO E, SERRANO-HERAS G, CASTAÑO M J, et al. Real-time PCR detection chemistry [J]. Clinica Chimica Acta, 2015, 439: 231-250. doi: 10.1016/j.cca.2014.10.017 [13] 吴彬彬, 卢滇楠, 刘铮. 石油污染土壤生物修复过程中氮循环功能基因的动态检测 [J]. 环境科学, 2012, 33(6): 2068-2074. WU B B, LU D N, LIU Z. Dynamic changes in functional genes for nitrogen cycle during bioremediation of petroleum-contaminated soil [J]. Environmental Science, 2012, 33(6): 2068-2074(in Chinese).
[14] NITSCHE A, STEUER N, SCHMIDT C A, et al. Different real-time PCR formats compared for the quantitative detection of human Cytomegalovirus DNA [J]. Clinical Chemistry, 1999, 45(11): 1932-1937. doi: 10.1093/clinchem/45.11.1932 [15] 康凌宇, 倪忠, 武俊明, 等. 实时荧光定量PCR技术在生物饲料中的应用 [J]. 中国饲料, 2021(1): 82-88. KANG L Y, NI Z, WU J M, et al. Application of real-time fluorescent quantitative PCR in biological feed [J]. China Feed, 2021(1): 82-88(in Chinese).
[16] 王玉倩, 薛秀花. 实时荧光定量PCR技术研究进展及其应用 [J]. 生物学通报, 2016, 51(2): 1-6. WANG Y Q, XUE X H. Research progress and application of real-time fluorescence quantitative PCR technology [J]. Bulletin of Biology, 2016, 51(2): 1-6(in Chinese).
[17] 刘庆龙, 唐景春, 万晓彤. 污染土壤中主要石油降解基因AlkB和Nah定量检测方法的建立和应用 [J]. 分析化学, 2014, 42(9): 1348-1353. doi: 10.11895/j.issn.0253-3820.140103 LIU Q L, TANG J C, WAN X T. Establishment of analysis method for detection of petroleum degrading genes AlkB and nah in contaminated soil and its application [J]. Chinese Journal of Analytical Chemistry, 2014, 42(9): 1348-1353(in Chinese). doi: 10.11895/j.issn.0253-3820.140103
[18] 宋本如, 甄梅楠, 刘小妹, 等. 石油烃厌氧降解基因masD和bamA实时荧光定量PCR方法的建立及应用 [J]. 分析化学, 2019, 47(2): 207-213. SONG B R, ZHEN M N, LIU X M, et al. Real-time fluorescence quantitative polymerase chain reaction for anaerobic degradation genes masD and BamA in petroleum hydrocarbons [J]. Chinese Journal of Analytical Chemistry, 2019, 47(2): 207-213(in Chinese).
[19] STAATS M, BRASTER M, RÖLING W F M. Molecular diversity and distribution of aromatic hydrocarbon-degrading anaerobes across a landfill leachate plume [J]. Environmental Microbiology, 2011, 13(5): 1216-1227. doi: 10.1111/j.1462-2920.2010.02421.x [20] KOLUKIRIK M, INCE O, INCE B K. Increment in anaerobic hydrocarbon degradation activity of halic bay sediments via nutrient amendment [J]. Microbial Ecology, 2011, 61(4): 871-884. doi: 10.1007/s00248-011-9825-8 [21] 李小康, 鱼涛, 李红, 等. 石油降解菌的复配及降解效果评价 [J]. 油田化学, 2020, 37(2): 311-317,324. LI X K, YU T, LI H, et al. Analysis of compounding and degradation of petroleum degrading bacteria [J]. Oilfield Chemistry, 2020, 37(2): 311-317,324(in Chinese).
[22] 苏世博, 蒲路莎, 陈肖韩, 等. 鹅源星状病毒TaqMan荧光定量PCR检测方法的建立及初步应用 [J]. 中国比较医学杂志, 2021, 31(3): 43-48. doi: 10.3969/j.issn.1671-7856.2021.03.007 SU S B, PU L S, CHEN X H, et al. Establishment and application of TaqMan reverse transcriptionquantitative PCR for newly emerging goose-origin astrovirus in China [J]. Chinese Journal of Comparative Medicine, 2021, 31(3): 43-48(in Chinese). doi: 10.3969/j.issn.1671-7856.2021.03.007
[23] 梁子英, 刘芳. 实时荧光定量PCR技术及其应用研究进展 [J]. 现代农业科技, 2020(6): 1-3,8. doi: 10.3969/j.issn.1007-5739.2020.06.001 LIANG Z Y, LIU F. Research progress on real-time quantitative PCR technology and its application [J]. Modern Agricultural Science and Technology, 2020(6): 1-3,8(in Chinese). doi: 10.3969/j.issn.1007-5739.2020.06.001
[24] 赵丽, 崔保安, 陈红英, 等. 鉴别伪狂犬病病毒野毒与疫苗毒荧光定量PCR方法的建立 [J]. 生物工程学报, 2008, 24(7): 1149-1154. doi: 10.3321/j.issn:1000-3061.2008.07.005 ZHAO L, CUI B A, CHEN H Y, et al. Diagonsis establishment of fluorescen quantitative PCR assay for pseudorabies wild-type virus and vaccine virus [J]. Chinese Journal of Biotechnology, 2008, 24(7): 1149-1154(in Chinese). doi: 10.3321/j.issn:1000-3061.2008.07.005
-



 下载:
下载: