-
多环芳烃(polycyclic aromatic hydrocarbons, PAHs)作为环境中普遍存在的一种典型持久性有机污染物,由2个或2个以上的苯环以直线、角状排列或聚簇状的方式构成[1]。PAHs因具有溶解度低、疏水性高、辛醇-水分配系数大以及难降解等特性[2],容易在环境中积累并在生物体内富集,对生物造成致畸、致癌、致突变的“三致效应”,且会通过食物链传递对人类健康造成危害[3]。
PAHs是指没有任何取代基和杂环原子的芳烃化合物[4],一般称为母体PAHs(parent PAHs, P-PAHs)。环境中还存在大量带有烷基等取代基的PAHs衍生物,其中烷基化多环芳烃(alkylated PAHs, A-PAHs)是以PAHs为母环,具有烷基侧链的稠环芳香烃,它在环境中主要来源于溢油[5-6]、煤燃烧[7]、生物质燃烧和尾气排放[8-9]。由成岩作用生成的A-PAHs是石油中总多环芳烃(T-PAHs,包括P-PAHs与PAHs衍生物)的主要形式,约占T-PAHs的85%—95%;泄漏到水环境后,沉积物中A-PAHs含量仍高达70%以上[10-11],其中以烷基萘、烷基菲为主[12]。烷基的取代会导致A-PAHs的水溶性比P-PAHs低,使其更倾向于在生物体内累积,产生比P-PAHs更大的毒性[13],在环境中很难被去除。
本文在实验室前期研究的基础上,结合国内外对A-PAHs的污染状况和生态毒性的研究现状,对细菌降解A-PAHs的代谢途径及关键降解基因和酶的研究进行了总结,有助于了解环境中A-PAHs的降解去除研究进展,为寻找高效的A-PAHs降解方法提供理论依据。
-
由于大气沉降、城市径流和水-气交换等作用,A-PAHs广泛分布于大气、土壤、水体等不同环境介质中[6,14-15]。Lian等[16]对2014年12月至2015年10月上海市降尘样品的季节与空间分布特征进行分析,发现41种A-PAHs的浓度范围为1.6—9.0 mg·g−1,其中烷基萘和烷基菲约占A-PAHs总含量的50%,且春冬两季中A-PAHs的浓度要高于夏秋两季;在空间分布上,上海西部地区的A-PAHs浓度高于东部地区。瑞典斯德哥摩尔幼儿园室内灰尘中PAHs含量最高的是菲及甲基菲衍生物,甲基菲衍生物的平均浓度约为1.3 pg·m−3[8]。Saha等[17]收集了印度、印度尼西亚、马来西亚等国家174个地区的表层沉积物样本,发现大多亚洲热带城市沉积物中T-PAHs均以A-PAHs为主,且城市地区的T-PAHs浓度远高于农村地区。
在原油泄露的环境,A-PAHs为T-PAHs的主要成分[18]。2007年“河北精神”号邮轮发生意外撞击,约1.1万吨原油泄漏入海,造成海域污染的3种货油中T-PAHs的浓度范围为4.64—8.50 mg·g−1,其中A-PAHs浓度约为P-PAHs的30倍[19]。在大连沿海地区发生溢油事故50 d后,在污染水体中能检测到25种A-PAHs,浓度范围为12.5—79.8 ng·L−1,约占T-PAHs含量的84.1%[20]。
我们课题组对珠江口、大亚湾和南海北部的表层沉积物中31种P-PAHs和29种A-PAHs进行了全面的分析调查,结果表明,A-PAHs在珠江口、大亚湾和南海北部的浓度范围分别为115—766、127—354、200—272 ng·g−1,占60种T-PAHs的32%—36%,在各A-PAHs中,浓度最高的为烷基萘和烷基菲[15]。综上所述,A-PAHs在全球分布广泛且存在于各种环境介质中,在原油污染环境中A-PAHs的浓度远高于P-PAHs,应对A-PAHs予以更多的关注。
-
在以往的研究中,针对PAHs环境效应及毒性作用机制的研究主要集中在16种优先控制的P-PAHs。而近年有研究表明,部分A-PAHs对鱼类早期发育的毒性强于P-PAHs。Turcotte等[21]通过日本青鳉(Oryzias latipes)的形态研究,比较了菲及其烷基化同系物的毒性,发现菲、1-甲基菲和2,7-二甲基菲的半数有效浓度(EC50)分别为95、76、35 μg·L−1,烷基菲对青鳉胚胎的毒性大于母体菲。在虹鳟鱼(Oncorhynchus Mykiss)的生长初期,惹烯(7-异丙基-1-甲基菲)的长期暴露会引起鱼类蓝囊病(Blue Sac Disease, BSD)、生长速率减慢[22]和细胞色素酶P450(CYP1A)的诱导反应[23]。诱导反应是惹烯与肝脏内的芳烃受体(aryl hydrocarbon receptors,AHRs)蛋白结合,诱导产生CYP1A蛋白[24],CYP1A催化PAHs的氧化反应,可能会产生活性氧和其它活性副产物,也可能诱导BSD的发生[25]。Mu等[10]发现菲和惹烯均可导致海洋青鳉(Oryzias melastigma)的胚胎发育畸形,其中惹烯对海洋青鳉早期发育的致死和致畸效应都明显强于其母体菲。毒理学研究表明,A-PAHs除了具有较强的“三致效应”外,还具有神经毒性,Chen等[26]发现3-甲基菲对斑马鱼单独暴露会导致其神经行为异常,与阻燃剂得克隆(dechlorane plus, DP)共同暴露会诱导其神经行为缺陷,使神经毒性更强。生物体受A-PAHs影响主要表现在生理活动紊乱、生理代谢异常、组织形状及功能发生变异等,甚至会引起繁殖能力异常,进而改变生物群落结构[27]。有研究表明,在受石油污染的环境中,水生生物体内的A-PAHs含量远远高于P-PAHs,A-PAHs可能是溢油污染中引起水生生物毒性的主要物质[28]。
除了对动物具有生态毒性,A-PAHs对植物也有毒性。植物会通过根、叶等对土壤和空气中的A-PAHs进行吸收而使正常生长受到影响,并在体内积累[29],对人类健康产生危害。Zhang等[30]研究了芘和1-甲基芘在玉米幼苗的生物积累和转运,不仅在玉米根中检测到这两种化学物质,而且发现它们可以转移到玉米幼苗的可食用部位。Sun等[31]通过研究玉米、大豆和马铃薯的3种作物叶片表面对菲、3-甲基菲、惹烯的吸收情况,发现3种PAHs的平衡吸附浓度随着芳香环上烷基取代的数量增加而增大。生物量、发芽率、色素含量和酶活性等指标可用来评价P-PAHs的植物毒性[32-35]。
目前对A-PAHs的生物毒性、在生物体内的代谢途径及毒理机制研究甚少。研究指出,A-PAHs的毒性效应受到烷基取代位点的影响,大多数甲基取代A-PAHs和其P-PAHs的致癌性差别不大,但有些甲基取代A-PAHs却明显不同于P-PAHs,如5-(或11-)甲基䓛表现出较强的致癌活性,6-(或12-)甲基䓛致癌活性稍强,而其他取代位置上的甲基䓛表现出较小的致癌活性[36]。早年,Jerina等[37]提出PAHs湾区理论的致癌机理,认为“最终”的致癌剂——二醇环氧化物环氧环打开后会生成与芳环共轭的碳正离子。Poulsen等[38]研究证实了湾区二醇环氧化物反应活性较高是由于其容易形成三醇碳正离子。通常认为三醇碳正离子的稳定性越高,其甲基取代PAHs的致癌性越强;离湾区三价碳正离子较近位置上的甲基取代PAHs具有异常高的致癌活性,这是甲基与碳正离子空间相互作用的结果[36]。另外,A-PAHs的毒性机制可能与在细胞色素酶P450作用下被快速转化为具有广泛结构和潜在毒性的代谢物有关[39]。A-PAHs的代谢中间产物,如多环芳香酸类[11]、5-甲基䓛-四醇、邻单磺化-5-甲基-儿茶酚[40]等物质,可能会引起更强的生态毒性。但是A-PAHs中间代谢产物的生态毒性效应的研究还比较缺乏。
-
微生物降解是A-PAHs从环境中降解去除的主要方法[2],与物理法、化学法相比,具有处理成本低、操作方便、没有二次污染、便于原位修复等特点。目前已经分离到许多能够降解A-PAHs的微生物,包括细菌、真菌和微藻。其中真菌中的雅致小克银汉霉(Cunninghamella elegans)能降解1-,2-甲基萘,其主要中间代谢物为反式1,2-,3,4-二氢二醇[41],黄孢原毛平革菌(Phanerochaete chrysosporiu)能够产生木质素过氧化物酶氧化1-,2-,和9-甲基蒽[42],茶树菇(Agrocybe aegerita)和辐毛小鬼伞(Coprinellus radians)产生的两种芳香过氧化物酶可催化氧化1-,2-甲基萘[43]。关于微藻对A-PAHs的降解研究非常少,目前只有我们团队的研究发现3种绿藻:近头状尖胞藻(Pseudokirchneriella subcapitata)、小球藻(Chlorella vulgaris)和斜生栅藻(Scenedesmus obliquus)对6种甲基菲(1-,2-,3-,4-,9-甲基菲和3,6-二甲基菲)具有不同的降解能力,近头状尖胞藻对A-PAHs的降解效率最高,7 d内对1-甲基菲的去除率为99.8%,对3,6-二甲基菲的去除率为75.6%,代谢产物均为单羟基甲基菲[44]。总的来说,真菌主要是利用自身产生的过氧化物酶对A-PAHs进行代谢,而藻类则通过加氧酶对A-PAHs进行氧化,与P-PAHs的降解机理一致。
影响微生物降解速率的因素主要有环境因素(温度、pH和金属离子等)、微生物种类及污染物物理化学性质等[45]。对A-PAHs来说,烷基取代基产生的空间位阻效应会使微生物的降解能力降低,还可能阻碍污染物进入细胞的跨膜过程[46-47]。空间位阻效应是指分子中某些原子或基团彼此接近而引起的空间阻碍作用[48]。Volkman等[49]对烷基萘的生物降解敏感性进行研究,发现烷基萘的生物降解速率从高到低为2-甲基萘 > 1-甲基萘 > 2,7-、1,7-和1,6-二甲基萘 > 1,3-和2,6-二甲基萘 > 1,4-和1,5-二甲基萘 > 1,2-二甲基萘 > 2,3-二甲基萘和乙基萘 > 2-乙基萘 > 1,8-二甲基萘。Huang等[50]指出烷基萘的生物降解过程与热力学降解过程不同,热力学稳定的异构体更容易被生物降解;A-PAHs不同异构体之间的生物降解存在显著差异性,可能与A-PAHs异构体分子结构有关,内能低且结构紧密的异构体相对容易被生物降解。Malmquist等[11]研究了沙蚕(Nereis diversicolor)对菲与甲基菲的降解过程及代谢产物,发现甲基菲代谢产物中的多环芳香酸(polycyclic aromatic acids, PAAs)含量高于菲代谢产生的PAAs含量,且这与甲基取代位置相关。总的来说,A-PAHs的生物降解规律与其化学结构有紧密联系,主要受芳烃化程度及烷基化程度两个因素影响,如果芳香烃中含有相邻的甲基取代基,生物降解速率会更慢,普遍趋势为β位甲基取代PAHs比α位甲基取代PAHs更容易发生降解[45,49]。
与真菌、藻类等相比,细菌降解A-PAHs的研究更多且大多降解菌都是从石油污染区域分离出来,较常见的有假单胞菌属(Pseudomonas)[51-53]、鞘氨醇菌属(Sphingomonads)[54-57]、微菌属(Micrococcus)[58]和分枝杆菌属(Mycobacterium)[56]。目前的研究主要集中在2—3环的A-PAHs。下面以甲基萘和甲基菲为代表,详细介绍细菌对A-PAHs的降解代谢途径。
-
萘(Naphthalene)是分子量最小的PAHs,因其具有结构简单且易从环境中分离得到降解菌株等原因,常被用作PAHs生物降解的模式化合物[59-61]。萘和甲基萘普遍存在于沉积有机质和原油中[62]。在过去几十年里,已经报道的甲基萘降解菌主要有假单胞菌属(Pseudomonas)[51-53]、鞘氨醇单胞菌属(Sphingomonas)[54]、解环菌属(Cycloclasticus)[63]和分枝杆菌属(Mycobacterium)[64]等,见表1。
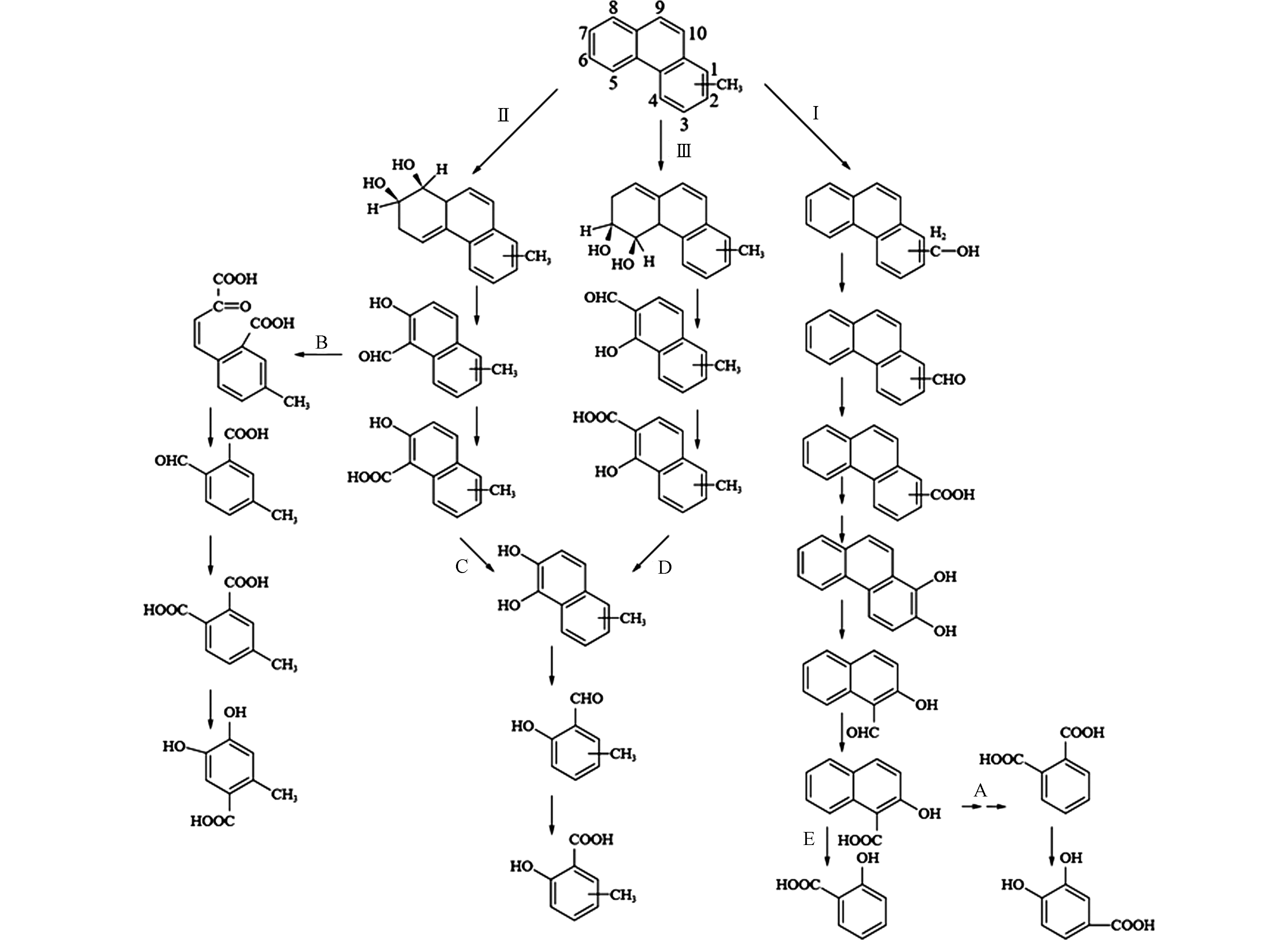
Kasai等[63]从原油污染海水中分离出的解环菌Cycloclasticus可在28 d内降解原油中80%以上的C0-4-烷基萘(C0表示未取代芳香族化合物,Cn中的数字表示侧链中的碳数)。向廷生等[67]利用气相色谱-质谱联用仪(GC-MS)分析技术,对辽河冷家油田轻中度生物降解程度原油中的烷基萘进行定性定量分析,发现烷基萘的生物降解性从易到难排序为:萘 > 甲基萘 > 乙基萘 > 二甲基萘 > 三甲基萘 > 四甲基萘,这表明A-PAHs的生物降解性会受到烷基化程度的影响。烷基萘的生物降解研究主要集中在甲基萘,特别是2-甲基萘。1964年,Davies和Evans[61]从土壤中分离出第一株对萘具有降解能力的假单胞菌Pseudomonas PG,并提出了萘的主要代谢途径。Williams等[51]通过更详细的酶学数据证实了这一途径,并表明Pseudomonas PG降解2-甲基萘时,菌株是以相同的途径——间位途径进行代谢。Mahajan等[52]从土壤中分离出一株可降解1-甲基萘和2-甲基萘的恶臭假单胞菌(Pseudomonas putida CSV86)且发现了甲基萘的多种代谢途径:一是在甲基端的氧化途径,甲基被羟基化生成羟甲基萘,然后羟甲基萘被羟甲基萘脱氢酶和萘甲醛脱氢酶催化转化为萘甲酸(图1-Ⅰ);二是未被甲基取代的芳香环羟基化为甲基邻苯二酚,随后在异构酶、水杨酸酶-醛缩酶和水杨醛脱氢酶的催化用下转化为甲基水杨酸,甲基水杨酸通过甲基邻苯二酚进入间位裂解途径进行下一步代谢,类似于萘的降解方式(图1-Ⅱ);三是未被甲基取代的芳香环和甲基可同时发生羟基化反应,随后产生4-羟甲基邻苯二酚再进入间位裂解途径进行代谢(图1-Ⅲ)。
以上为一个甲基取代的情况,对于多个甲基取代基,Dutta等[54]用Sphingomonas paucimobilis 2322对2-甲基萘、2,6-、2,7-、2,3-、1,5-、1,6-、1,8-、1,2-、1,4-二甲基萘和2,3,6-三甲基萘等多种甲基萘进行了进一步的降解研究,提出了S. paucimobilis 2322对多种甲基萘的转化路径:当甲基分布于两个芳香环时,首先对其中一个甲基进行氧化,生成-COOH基团再进入下一步开环过程或另一个甲基的氧化过程(图1-Ⅳ);当甲基存在于同一个芳香环时,有两条降解途径:一是非甲基一侧芳香环的氧化开环,二是甲基端氧化生成终产物-COOH(图1-Ⅲ、Ⅴ)。从图1可以看出,烷基的取代及烷基化程度的增加,丰富了甲基萘的降解途径,但是只有未被烷基取代的芳香环上的双加氧氧化才能使A-PAHs进一步开环,降解成为分子量更小的化合物(Ⅱ和Ⅲ路径)。烷基取代基上面的单加氧氧化难以让化合物进一步开环降解(Ⅰ、Ⅳ和Ⅴ路径),会造成代谢产物的积累,不利于化合物的矿化,这也可能是A-PAHs的降解速率小于其P-PAHs的原因。
此外,研究还发现2-甲基萘可在硫酸盐还原[68]和产甲烷条件[69]的情况下发生厌氧生物降解。Folwell等[70]研究表明,2-甲基萘可在上述条件下被降解,生成2-萘甲酸为中间代谢产物;变性梯度凝胶电泳(DGGE)结果表明,2-甲基萘在厌氧生物降解过程中起主要降解作用的菌属是Fusibacter,Alkaliphilus,Desulfobacterium,Variovorax,Thaurea和Hydrogenophaga。
-
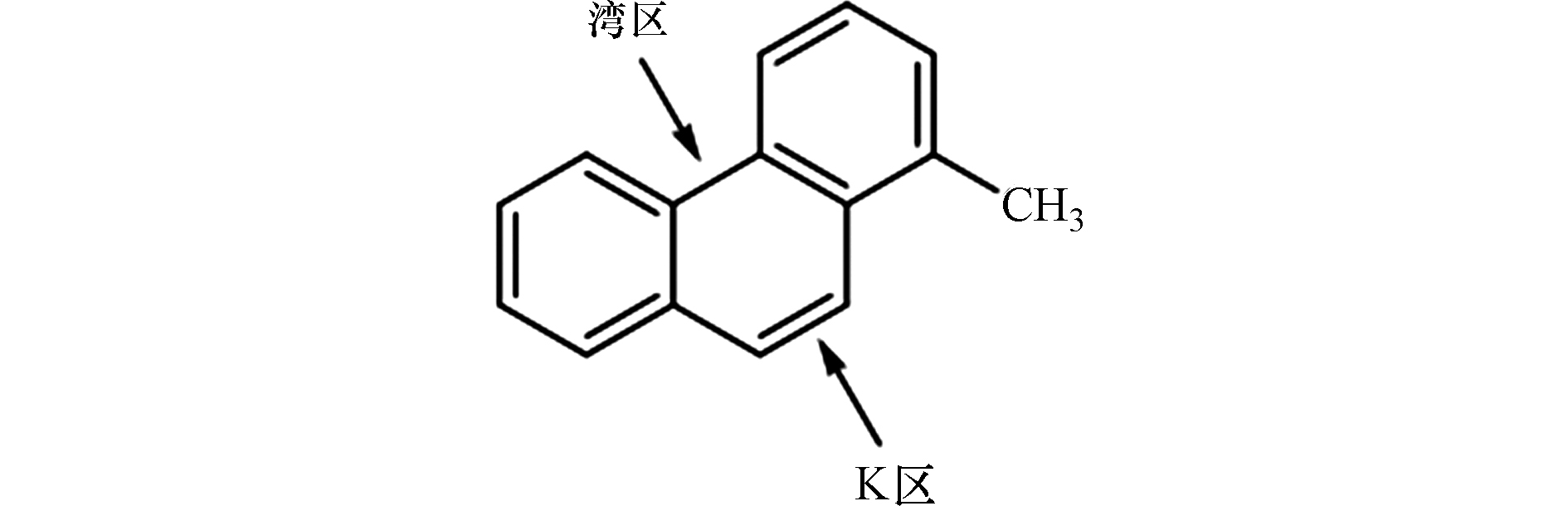
烷基菲是石油中最常见、含量最丰富的一种A-PAHs,主要来源于沉积盆地中沉积有机质的成岩作用、化石或生物质的燃烧,其浓度和分布受初级有机输入和次级改造过程的控制[71-72]。因烷基菲是同时含有“湾区”和“K区”的分子量最小的A-PAHs(图2),在研究中常被用作A-PAHs的模式化合物,并作为检测A-PAHs污染的标志性化合物[71,73]。
烷基菲的细菌降解研究主要集中在甲基菲。目前已知能降解甲基菲的菌属有分枝杆菌属(Mycobacterium)、鞘氨醇单胞菌属(Sphingomonas)、微球菌属(Micrococcus)和海迪茨氏菌(Dietzia maris)等,这些菌属对母体菲同样具有降解效果[56-58,74-75],见表2。
鞘氨醇单胞菌属对多种A-PAHs都有很好的降解能力,表现出明显的代谢多样性。Gilewicz等[74]从炼油厂附近受污染的土壤中分离出一株可以2-甲基菲为单一碳源和能源的菌株Sphingomonas sp. 2MPII,可将初始浓度为200 mg·L−1的2-甲基菲在5 d内降解70%。Sabate等[75]从杂酚油污染土壤中筛选出另一株可降解2-甲基菲的菌株Sphingomonas sp. JS5,可在3 d内将初始浓度为86.4 mg·L−1的2-甲基菲降至2.7 mg·L−1。我们课题组从广州石化厂原油污染土壤中筛选出一株对1-甲基菲具有降解能力的新型革兰氏阴性菌Novosphingobium guangzhouense sp. Nov.,该菌株对100 mg·L−11-甲基菲的10 d降解率大于80%[76]。Wang等[77]发现,无色杆菌Achromobacter strain J3和柠檬酸杆菌Citrobacter sp. strain J1对甲基菲具有较好的生物降解作用,添加表面活性素(由不同的枯草芽孢杆菌菌株产生的一种环状脂肽)可使其生物降解率提高1.5%—87.2%。
烷基菲的烷基取代数目会影响烷基菲的生物可利用性,烷基取代数目越多越难以被生物降解,生物降解难易程度从易到难的顺序为:菲 > 甲基菲 > C2-菲 > C3-菲(C2表示有2个碳数取代,包括二甲基和乙基;C3表示有3个碳数取代的组合)[49]。甲基取代位点的不同会给甲基菲的生物降解性带来一定的差异,9-甲基菲最难被生物降解,3-甲基菲、2-甲基菲和1-甲基菲的可生物降解性相当[67,78-79]。
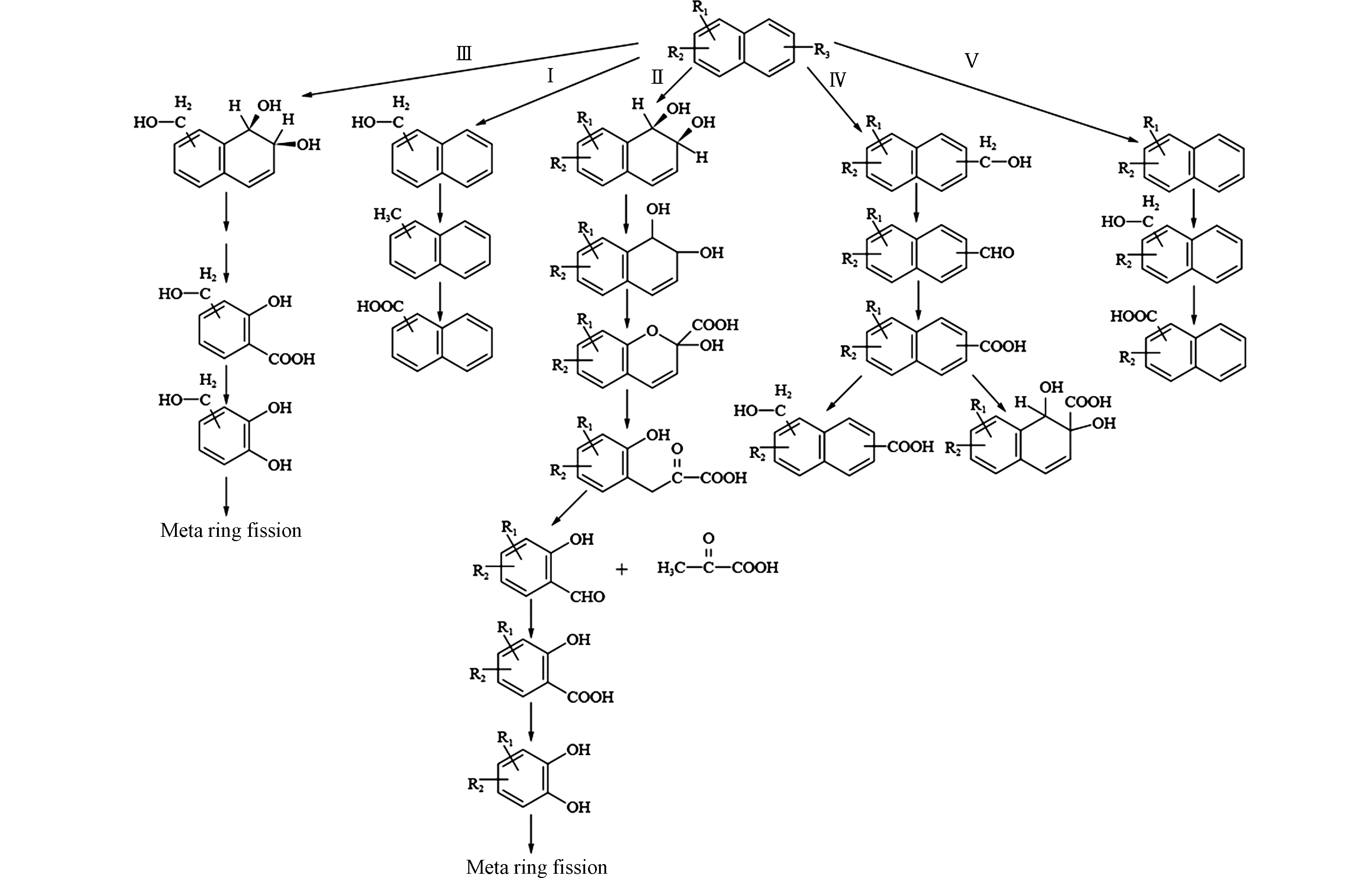
Budzinski等[80]从地中海沿岸沉积物中分离出一株可降解2-甲基菲的菌株Sphingomonas sp. 2MPII,通过GC-MS分析鉴定降解过程中产生的代谢产物,发现2-甲基菲有两种不同的代谢途径:一条是从甲基开始氧化,生成2-菲甲酸,然后氧化脱羧,生成1,2-二羟基菲,进入菲的降解途径(图3-Ⅰ-A);另一条是在非甲基端芳香环上进行双加氧反应,从远到近逐个对芳香环进行开环,最后沿着两条支路分别生成4-甲基邻苯二酸(图3-Ⅱ-B)和4-甲基水杨酸(图3-Ⅱ-C)[59]。
我们团队的钟佳南等[57]从石油污染土壤中分离出的Sphingobium sp. MP9-4菌株可降解1-甲基菲,通过代谢产物的分析鉴定,发现其代谢途径与Budzinski等[80]的研究结果相似,一条途径是分别在芳香环的C-7,8和C-5,6位置进行双加氧反应,分别开环形成2-羟基-5-甲基-1-萘醛和1-羟基-5-甲基-2-萘醛,各自氧化成相应的酸,经过脱羧,均生成5-甲基-1,2-二萘酚,进入共同的降解通路(图3-Ⅱ-C、3-Ⅲ-D);另一条途径为甲基端的氧化,可以生成标志性的1-菲甲酸,脱羧氧化形成1,2-二羟基菲,由此进入菲的双加氧降解通路,逐步生成2-羟基-1-萘醛、2-羟基-1-萘酸、水杨酸等(图3-Ⅰ-E)。
综上所述,甲基菲的降解途径主要以芳香环处双加氧反应和甲基端单加氧反应为主,与甲基萘的降解类似。由于菌株或降解条件的不同,在降解过程中往往会出现不同的中间代谢产物。一个菌株通常不会只有一条降解途径,它可能同时拥有两条及以上的代谢途径,并根据环境的条件来决定降解酶促反应的方向。在A-PAHs细菌降解过程中,由于其与P-PAHs的结构差异,烷基侧链一般很难被酶去除,这使得A-PAHs的生物可利用性比P-PAHs更低,从而增大了降解难度[58]。
-
PAHs细菌降解的初始步骤是加氧酶的氧化作用,加氧酶包括单加氧酶和双加氧酶,其中以双加氧酶的作用为主。Kelley等[81]对一株分枝杆菌(Mycobacterium sp.)降解萘的初步代谢路径研究发现,菌株最初分别通过双加氧酶和单加氧酶催化反应转化为顺式-和反式-1,2-二羟基-1,2-二氢化萘,顺式和反式产物的比例约为25:1;顺式产物由双加氧酶途径生成,反式产物由单加氧酶途径生成,说明细菌对PAHs的降解以双加氧酶途径为主。从上文细菌对A-PAHs降解途径的报道可知,其降解途径可通过单加氧酶从烷基端开始氧化,也可由双加氧酶对芳香环进行初始氧化。但目前对A-PAHs降解酶与降解基因的研究几乎处于空白状态,A-PAHs芳香环上的氧化可参考P-PAHs的氧化通路,烷基端的氧化则以带有烷基的芳香烃的氧化通路作为参考。
-
双加氧酶根据开环时起作用的前后顺序可分为两种,一种是环羟化双加氧酶,负责催化氧对芳香环的进攻,同时将两个氧原子引入苯环,直接生成带有双羟基的顺式二氢二醇;另一种是环裂解双加氧酶,负责催化被氧化后的芳香环的开裂[82]。环羟化双加氧酶的代表有甲苯双加氧酶、联苯双加氧酶和萘双加氧酶[83-84]。环裂解双加氧酶又分为两种,催化邻位开环的外断裂开环酶,代表为邻苯二酚-1,2-双加氧酶,和催化间位开环的内断裂开环酶,代表为邻苯二酚-2,3-双加氧酶[85-86]。Tondervik等[87]利用重组表达的Ralstonia sp. U2菌株的萘双加氧酶(NDO)和Comamonas sp. JS765菌株的硝基苯双加氧酶(NBDO)对大量甲基萘进行实验,发现NDO和NBDO对甲基萘既能进行单加氧反应,也能进行双加氧反应,其中,在大多降解过程中NBDO的双加氧产物水平高于单加氧产物水平。在Sphingobium sp. MP9-4对甲基菲的降解过程中,我们也发现双加氧产物的生成量远高于单加氧产物的含量[57]。
多种功能酶在PAHs代谢过程中被发现,如羟基化双加氧酶、脱氢酶和水合酶-醛缩酶等[88],但大多研究都是围绕低分子量PAHs降解基因进行[89]。PAHs的降解从羟基化双加氧酶对苯环的羟基化开始,该酶是降解过程中的关键酶和限速酶,且是一个多组分的复杂酶系[90]。这个酶系一般由三个部分组成:黄素蛋白、铁硫蛋白和铁氧还蛋白[91-92]。通过对双加氧酶基因结构和功能的研究,许多降解菌株的相关降解基因或基因簇被发现[93]。
以同源性关系为依据对PAHs降解基因进行分类,nah-like基因占很大一部分,这是一类PAHs降解通路的上游基因。例如假单胞菌Pseudomonas putida G7的NAH7质粒中nah基因的基因簇排列顺序为nahAaAbAcAdBFCQED[94],菌株降解过程中的不同酶系由nah-like中不同位点的基因分别编码,如萘羟基化双加氧酶(NDO)的铁还原蛋白酶由nahAa基因编码,NDO的铁还原蛋白由nahAb基因编码,NDO大亚基由nahAc基因编码等[95]。与经典的nah基因相似的nag基因,在罗尔斯通菌Ralstonia sp. U2中的基因簇排列顺序为nagAaGHAbAcAdBFCQED,除了nagGH基因以外,基因簇与nah基因顺序相同,功能相近[87]。另有一类基因的基因簇顺序和同源性均与nah-like基因不同,为phd基因。Zylstra等[96]报道了丛毛单胞菌Comamonas testosteroni GZ39降解菲的phd基因顺序为phdAbAaBAcAdDE,phdAa基因编码铁氧化还原蛋白还原酶,phdAb基因编码铁氧化还原蛋白,phdAc和phdAd基因分别编码铁硫蛋白的大小亚基,phdB、phdD和phdE基因分别编码顺式-二羟基脱氢酶、异构酶和水合酶-醛缩酶。在部分革兰氏阳性菌中也存在phd基因,Saito等[97]发现Nocardioides sp. strain KP7编码降解菲的基因簇phdEFABGHCD,phdA和phdB基因编码菲双加氧酶的大小亚基,phdC基因编码铁氧化还原蛋白,phdD基因编码铁氧化还原蛋白还原酶。Laurie等[98]研究了菌株Burkholderia sp. RP007中phn基因对菲的降解,发现phn基因虽在功能上与nah-like基因相似,但二者同源性非常低[99],其基因顺序为phnRSFECDAcAdB,与phd基因功能相似,phnAc和phnAd基因分别编码起始双加氧酶铁硫蛋白的大小亚基,phnB、phnD和phnE基因分别编码二羟基脱氢酶、异构酶和水合酶-醛缩酶,另外phnC基因编码外源型双加氧酶,phnF基因编码乙醛脱氢酶。
此外,PAHs的降解基因还有nar和nid基因。Rhodococcus是一种经典的可降解PAHs的革兰氏阳性菌,具有强大的代谢功能,与革兰氏阴性菌对萘的降解基因不同的是,它只有3个萘降解所需的结构基因narAa、narAb和narB,narAa和narAb基因分别编码NDO的大小亚基[100-101]。Kumari等[102]从石油污染场地分离出一株PAHs降解菌Stenotrophomonas sp. IITR87,以萘双加氧酶基因(nidA)为探针,对菌株IITR87进行Southern杂交,发现了nidA基因的存在,PCR分析表明nid基因分散出现在基因组中,不同于芘降解菌Mycobacterium vanbaalenii PYR-1中成簇出现[103],菌株IITR87的nid基因中nidA和nidB基因分别编码萘双加氧酶的大小亚基,nidD基因编码醛脱氢酶,与已报道的Mycobacterium vanbaalenii PYR-1中的nid基因的同源性均超过97%。环境中还存在一些其他类型的降解基因簇,如pht[103]和pdo[104-105]等。
从A-PAHs的降解途径来看,A-PAHs未被烷基取代的芳香环上的氧化反应与其P-PAHs完全一致,可以推测上述提到的芳香环氧化降解酶与基因也会存在于A-PAHs降解菌上,原因是目前发现的A-PAHs降解菌对其P-PAHs同样具有降解效果,它们很可能共用一套酶系。随着对PAHs微生物降解在分子水平认识的不断深入,能够确定的降解基因数目越来越多,功能越来越明确,这些研究成果都将为A-PAHs降解基因和降解酶的研究提供更多的参考依据。
-
A-PAHs在烷基上的氧化通路可参考烷基苯,例如甲苯、二甲苯等。甲苯和二甲苯从甲基开始的降解过程基本相同,首先将甲基逐步氧化成-CH2OH、-CHO和-COOH,接着脱羧形成双羟基苯,进行开环裂解,再经过一些步骤,最后进入三羧酸循环[106]。研究表明,甲苯和二甲苯的降解基因多集中在同一条质粒上,例如,pWW0 TOL质粒,主要编码降解甲苯、间二甲苯和邻二甲苯等所需要的酶,TOL质粒有上游操纵子和下游操纵子,分别负责甲苯和二甲苯甲基端的氧化和进一步的开环降解,上游操纵子上有7个降解酶基因,下游操纵子有13个降解酶基因和2个调节基因[107],它们编码的蛋白中,可能与A-PAHs的烷基端降解相关的酶有甲基氧化酶XylM及其电子传递亚基XylA、苯甲醇脱氢酶XylB、苯甲醛脱氢酶XylC、苯甲酸双加氧酶的氧化酶α亚基XylX、β亚基XylY和还原酶XylZ、脱羧酶XylL、铁氧还原蛋白酶XylT[108]。benABC基因编码苯甲酸双加氧酶,与XylXYZ功能相似[109];benD基因编码脱羧酶,与XylL基因功能相似[110]。在1-甲基-4-异丙基苯降解通路中发现了cymAaAbC的3个基因,其中cymAa和cymAb分别编码1-甲基-4-异丙基苯初始氧化步骤单加氧酶的铁硫蛋白和铁氧还原蛋白部分,将对异丙苯甲醛氧化成对异丙苯甲酸的芳香醛脱氢酶由cymC编码[111]。此外,芳香醇脱氢酶可将-CH2OH氧化成-CHO,该酶参与多种芳香化合物的氧化过程,包括甲苯、二甲苯、苯丙氨酸等[112]。
除了好氧降解以外,在甲苯厌氧降解过程中,甲苯上的甲基碳原子在苯甲基琥珀酸合成酶的作用下与延胡索酸根离子上的双键结合生成苯甲基琥珀酸,再进行一系列β氧化生成苯甲酰辅酶A[110]。在A-PAHs的厌氧降解中,Meckenstock等[113]研究发现2-甲基萘的甲基可通过添加富马酸被激活,生成萘基-2-甲基琥珀酸盐为主要代谢产物,此现象与反硝化细菌厌氧降解甲苯类似,表明萘-2-甲基琥珀酸合成酶反应也由甘氨酰自由基酶催化。Pérez-Jiménez等[114]分析了异化硫酸还原酶的功能基因dsrAB,发现该基因编码蛋白质的α和β亚基来催化硫酸盐还原的最后一步,作者利用能矿化石油组分中烷烃、甲苯等物质的dsrAB基因对硫酸盐还原菌(SRB)进行表征,以苯、萘、2-甲基萘和菲作为唯一碳源再进行矿化,克隆了22个新的dsrAB基因并测序,发现这些dsrAB基因广泛分布在已知的SRB和已报道的环境克隆dsrAB中,表明许多可生物降解的SRB在系统发育上是不同的。
由于A-PAHs上的芳香环双加氧反应与烷基端的单加氧反应是同时进行,因此A-PAHs降解菌株上芳香环氧化与烷基氧化的降解酶与降解基因是同时表达的,但目前还没有此方面的报道,值得进一步研究。
-
A-PAHs在环境中广泛存在,是原油污染环境中PAHs衍生物的主要成分。由于烷基取代基的增加,导致某些A-PAHs化学结构更加稳定,生态毒性也往往强于其P-PAHs。微生物降解是A-PAHs从环境中去除的最主要途径,以细菌降解研究最多,以好氧降解为主。目前的研究主要集中在2—3环的A-PAHs,以甲基萘和甲基菲为代表,降解菌种类与P-PAHs类似,以假单胞菌属、鞘氨醇菌属和分枝杆菌属为主,降解途径以未被烷基取代的芳香环双加氧反应和烷基端单加氧反应为主,烷基端的单加氧反应最终也会进入P-PAHs的双加氧降解途径,由此可以推测细菌降解A-PAHs与P-PAHs共用一套降解酶系,但是目前关于A-PAHs相关降解酶、降解基因等方面的研究内容还知之甚少,对A-PAHs降解基因与酶的研究可以参考P-PAHs的降解基因与酶。
为了探求更高效的A-PAHs的降解途径及达到生态友好修复的目的,需要更全面地了解A-PAHs在各个环境介质中的污染现状,深入认识A-PAHs的生物累积效应及规律,开展A-PAHs中间代谢产物的生态毒性效应研究,为其健康风险评价提供更全面的毒性数据。我们需从分子生物学方面更深入地了解细菌降解A-PAHs的全过程,深入探究相关降解基因和关键酶的功能,由此可通过基因重组构建A-PAHs高效降解菌株。至今,人们对于低分子量A-PAHs的微生物降解研究相对较多,高分子量A-PAHs降解路径的研究还有待深入开展,全面了解和完善A-PAHs的生物降解过程,为在实际环境中A-PAHs的污染修复提供理论依据。
烷基化多环芳烃的细菌降解研究进展
Research progress on bacterial degradation of alkylated polycyclic aromatic hydrocarbons
-
摘要: 烷基化多环芳烃(alkylated polycyclic aromatic hydrocarbons, A-PAHs)是以多环芳烃(PAHs)为母环,具有烷基侧链的稠环芳香烃,是一类在环境中广泛存在的持久性有机污染物。微生物降解是其在环境中降解去除的主要途径,与真菌、藻类等相比,细菌降解A-PAHs得到更多的关注。本文对A-PAHs的污染现状及生态毒性,细菌降解甲基萘、甲基菲的研究进展进行了概述,以PAHs的降解酶和降解基因作为参考,总结了A-PAHs可能涉及的降解酶及降解基因。本文有助于了解环境中A-PAHs的生物降解研究现状,为寻找高效的A-PAHs降解方法及减轻其生态风险提供理论依据。Abstract: Alkylated polycyclic aromatic hydrocarbons (A-PAHs) are polycyclic aromatic hydrocarbons (PAHs) with parent ring and alkyl side chain, which are a kind of toxic persistent organic pollutants that widely exist in the environment. Microbial degradation is the main way for their removal in the environment. Compared with fungi and algae, more studies have been focused on bacterial degradation. The pollution status and ecological toxicity of A-PAHs, as well as the research progress of bacterial degradation of methylnaphthalene and methylphenanthrene were summarized in this review. Taking the degradation enzymes and genes of PAHs as reference, the possible degradation enzymes and genes involved in A-PAHs degradation bacteria were summarized. It is helpful to understand the research progress of A-PAHs biodegradation in the environment and provide theoretical basis for finding the efficient degradation methods of A-PAHs and reducing their ecological risks.
-

-
表 1 甲基萘降解菌的来源、底物、底物强化浓度及降解速率
Table 1. Source, substrate, substrate enrich concentration and degradation rate of methylnaphthalene by bacteria strain
菌株名称
Bacteria strain来源
Source底物
Substrate底物强化浓度
Substrate enrich
concentration降解速率
Degradation rate参考文献
ReferencePseudomonas PG 土壤 萘、2-甲基萘 330 mg·L−1 — [51] Pseudomonas putida CSV86 土壤 1-甲基萘、2-甲基萘 0.15%(W/V) — [52] Pseudomonas sp. strain NGK1 土壤 2-甲基萘 0.2%(W/V) — [53] Sphingomonas paucimobilis 2322 杂酚油污染土壤 二甲基萘、2-甲基萘 1000 mg·L−1 — [54] Cycloclasticus 原油污染海水 C0-4-烷基萘 1000 mg·L−1 1.190 mg·L−1·h−1 [63] Mycobacterium sp. RJGII-135 土壤 1-甲基萘、2-甲基萘 (9—17)×10−3 mg·L−1 0.236×10−3 mg·L−1·h−1 [64] Neptunomonas naphthovorans
strain NAG-2N-113杂酚油污染沉积物 1-甲基萘、2-甲基萘 5 mg·L−1 0.010 mg·L−1·h−1 [65] Paenbacillus alvei 土壤 2-甲基萘 1.0%(V/V) 33% ,3 weeks [66] 表 2 甲基菲降解菌的来源、底物、底物强化浓度及降解速率
Table 2. Source, substrate, substrate enrich concentration and degradation rate of methylnaphthalene by bacteria strain
菌株名称
Bacteria strain来源
Source底物
Substrate底物强化浓度/
(mg·L−1)
Substrate enrich
concentration降解速率/
(mg·L−1·h−1)
Degradation rate参考文献
ReferenceSphingomonas sp. LH128 土壤 1-甲基菲 100 — [56] Sphingomonas subartica Arj13 土壤 1-甲基菲 100 (0.004 ± 0.001) [56] Sphingomonas sp. Arj19 土壤 1-甲基菲 100 (0.010 ± 0.001) [56] Sphingomonas sp. Arj81 土壤 1-甲基菲 100 (0.056 ± 0.008) [56] Sphingomonas sp. EPA505 土壤 1-甲基菲 100 (0.010 ± 0.001) [56] Mycobacterium gilvum RL1 土壤 2-甲基菲 100 (0.093 ± 0.007) [56] Mycobacterium aurum Ri464 土壤 2-甲基菲 100 (0.078 ± 0.012) [56] Mycobacterium austroafricanum Ri452-b 土壤 2-甲基菲 100 (0.130 ± 0.022) [56] Sphingomonas sp. MP9-4 石化原油污染土壤 1-甲基菲 10 0.104 [57] Micrococcus sp. CBMAI 636 地层水,坎波斯盆地 1-甲基菲、2-甲基菲、
3-甲基菲和9-甲基菲— 30 %, 21 d [58] Dietzia maris CBMAI705 石油,坎波斯盆地 1-甲基菲、2-甲基菲、
3-甲基菲和9-甲基菲— 99%, 21 d [58] Sphingomonas sp. 2MPII 炼油厂污染土壤 2-甲基菲 200 1.667 [74] Sphingomonas sp. JS5 杂酚油污染土壤 2-甲基菲 86.4 1.163 [75] Novosphingobium guangzhouense sp. Nov 石化原油污染土壤 1-甲基菲 100 0.417 [76] Achromobacter strain J3 石油污染土壤 甲基菲 — — [77] Citrobacter sp. strain J1 石油污染土壤 二甲基菲、
三甲基菲— — [77] -
[1] BAMFORTH S M, SINGLETON I. Bioremediation of polycyclic aromatic hydrocarbons: Current knowledge and future directions [J]. Journal of Chemical Technology & Biotechnology, 2005, 80(7): 723-736. [2] HARITASH A K, KAUSHIK C P. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): A review [J]. Journal of Hazardous Materials, 2009, 169(1/2/3): 1-15. [3] GHOSAL D, GHOSH S, DUTTA T K, et al. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): A review [J]. Frontiers in Microbiology, 2016, 7: 1369. [4] FETZER J C. The chemistry and analysis of large PAHs [J]. Polycyclic Aromatic Compounds, 2007, 27(2): 143-162. doi: 10.1080/10406630701268255 [5] NOH J, KIM H, LEE C, et al. Bioaccumulation of polycyclic aromatic hydrocarbons (PAHs) by the marine clam, Mactra veneriformis, chronically exposed to oil-suspended particulate matter aggregates [J]. Environmental Science & Technology, 2018, 52(14): 7910-7920. [6] HONG W J, JIA H L, YANG M, et al. Distribution, seasonal trends, and lung cancer risk of atmospheric polycyclic aromatic hydrocarbons in North China: A three-year case study in Dalian city [J]. Ecotoxicology and Environmental Safety, 2020, 196: 110526. doi: 10.1016/j.ecoenv.2020.110526 [7] HUANG Q, ZHU Y X, WU F, et al. Parent and alkylated polycyclic aromatic hydrocarbons in surface sediments of mangrove wetlands across Taiwan Strait, China: Characteristics, sources and ecological risk assessment [J]. Chemosphere, 2021, 265: 129168. doi: 10.1016/j.chemosphere.2020.129168 [8] LIM H, SADIKTSIS I, de OLIVEIRA GALVÃO M F, et al. Polycyclic aromatic compounds in particulate matter and indoor dust at preschools in Stockholm, Sweden: Occurrence, sources and genotoxic potential in vitro [J]. Science of the Total Environment, 2021, 755: 142709. doi: 10.1016/j.scitotenv.2020.142709 [9] CASAL C S, ARBILLA G, CORRÊA S M. Alkyl polycyclic aromatic hydrocarbons emissions in diesel/biodiesel exhaust [J]. Atmospheric Environment, 2014, 96: 107-116. doi: 10.1016/j.atmosenv.2014.07.028 [10] MU J L, WANG J Y, JIN F, et al. Comparative embryotoxicity of phenanthrene and alkyl-phenanthrene to marine medaka (Oryzias melastigma) [J]. Marine Pollution Bulletin, 2014, 85(2): 505-515. doi: 10.1016/j.marpolbul.2014.01.040 [11] MALMQUIST L M V, SELCK H, JØRGENSEN K B, et al. Polycyclic aromatic acids are primary metabolites of alkyl-PAHs—A case study with Nereis diversicolor [J]. Environmental Science & Technology, 2015, 49(9): 5713-5721. [12] YANG C, WANG Z D, HOLLEBONE B P, et al. Chromatographic fingerprinting analysis of crude oils and petroleum products[M]//Handbook of Oil Spill Science and Technology. Hoboken, NJ: John Wiley & Sons, Inc, 2015: 93-163. [13] FALLAHTAFTI S, RANTANEN T, BROWN R S, et al. Toxicity of hydroxylated alkyl-phenanthrenes to the early life stages of Japanese medaka (Oryzias latipes) [J]. Aquatic Toxicology, 2012, 106/107: 56-64. doi: 10.1016/j.aquatox.2011.10.007 [14] HINDERSMANN B, ACHTEN C. Urban soils impacted by tailings from coal mining: PAH source identification by 59 PAHs, BPCA and alkylated PAHs [J]. Environmental Pollution, 2018, 242: 1217-1225. doi: 10.1016/j.envpol.2018.08.014 [15] YUAN K, WANG X W, LIN L, et al. Characterizing the parent and alkyl polycyclic aromatic hydrocarbons in the Pearl River Estuary, Daya Bay and northern South China Sea: Influence of riverine input [J]. Environmental Pollution, 2015, 199: 66-72. doi: 10.1016/j.envpol.2015.01.017 [16] LIAN J J, REN Y, CHEN J M, et al. Distribution and source of alkyl polycyclic aromatic hydrocarbons in dustfall in Shanghai, China: The effect on the coastal area [J]. J Environ Monit, 2009, 11(1): 187-192. doi: 10.1039/B814232G [17] SAHA M H, TOGO A, MIZUKAWA K, et al. Sources of sedimentary PAHs in tropical Asian waters: Differentiation between pyrogenic and petrogenic sources by alkyl homolog abundance [J]. Marine Pollution Bulletin, 2009, 58(2): 189-200. doi: 10.1016/j.marpolbul.2008.04.049 [18] WAMMER K H, PETERS C A. Polycyclic aromatic hydrocarbon biodegradation rates: A structure-based study [J]. Environmental Science & Technology, 2005, 39(8): 2571-2578. [19] YIM U H, HA S Y, AN J G, et al. Fingerprint and weathering characteristics of stranded oils after the Hebei Spirit oil spill [J]. Journal of Hazardous Materials, 2011, 197: 60-69. doi: 10.1016/j.jhazmat.2011.09.055 [20] HONG W J, JIA H L, SUN Y Q, et al. Distribution, source and ecological risk assessment of parent and alkylated PAHs in coastal environment of Dalian, China after oil spill [J]. Polycyclic Aromatic Compounds, 2020, 40(4): 998-1012. doi: 10.1080/10406638.2018.1517809 [21] TURCOTTE D, AKHTAR P, BOWERMAN M, et al. Measuring the toxicity of alkyl-phenanthrenes to early life stages of medaka (Oryzias latipes) using partition-controlled delivery [J]. Environmental Toxicology and Chemistry, 2011, 30(2): 487-495. doi: 10.1002/etc.404 [22] BILLIARD S M, QUERBACH K, HODSON P V. Toxicity of retene to early life stages of two freshwater fish species [J]. Environmental Toxicology and Chemistry, 1999, 18(9): 2070-2077. doi: 10.1002/etc.5620180927 [23] BRINKWORTH L C, HODSON P V, TABASH S, et al. cyp1a induction and blue sac disease in early developmental stages of rainbow trout (Oncorhynchus mykiss) exposed to retene [J]. Journal of Toxicology and Environmental Health, Part A, 2003, 66(7): 627-646. doi: 10.1080/15287390309353771 [24] BILLIARD S M, HAHN M E, FRANKS D G, et al. Binding of polycyclic aromatic hydrocarbons (PAHs) to teleost aryl hydrocarbon receptors (AHRs) [J]. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 2002, 133(1): 55-68. doi: 10.1016/S1096-4959(02)00105-7 [25] HALLIWELL B, GUTTERIDGE J M C. Free radicals in biology and medicine, second edition [J]. Free Radical Biology and Medicine, 1991, 10(6): 449-450. doi: 10.1016/0891-5849(91)90055-8 [26] CHEN X P, CHEN Y H, HUANG C J, et al. Neurodevelopmental toxicity assessments of alkyl phenanthrene and Dechlorane Plus co-exposure in zebrafish [J]. Ecotoxicology and Environmental Safety, 2019, 180: 762-769. doi: 10.1016/j.ecoenv.2019.05.066 [27] FINGAS M. The basics of oil spill cleanup [J]. Journal of Fusion Energy, 2012, 12(1/2): 53-57. [28] NEFF J M, STOUT S A, GUNSTER D G. Ecological risk assessment of polycyclic aromatic hydrocarbons in sediments: Identifying sources and ecological hazard [J]. Integrated Environmental Assessment and Management, 2005, 1(1): 22-33. doi: 10.1897/IEAM_2004a-016.1 [29] ABDEL-SHAFY H I, MANSOUR M S M. A review on polycyclic aromatic hydrocarbons: Source, environmental impact, effect on human health and remediation [J]. Egyptian Journal of Petroleum, 2016, 25(1): 107-123. doi: 10.1016/j.ejpe.2015.03.011 [30] ZHANG H Y, LIU Y, SHEN X F, et al. Influence of multiwalled carbon nanotubes and sodium dodecyl benzene sulfonate on bioaccumulation and translocation of Pyrene and 1-methylpyrene in maize (Zea mays) seedlings [J]. Environmental Pollution, 2017, 220: 1409-1417. doi: 10.1016/j.envpol.2016.10.093 [31] SUN H F, GUO S, NAN Y L, et al. Direct determination of surfactant effects on the uptake of gaseous parent and alkylated PAHs by crop leaf surfaces [J]. Ecotoxicology and Environmental Safety, 2018, 154: 206-213. doi: 10.1016/j.ecoenv.2018.02.045 [32] SVERDRUP L E, KROGH P H, NIELSEN T, et al. Toxicity of eight polycyclic aromatic compounds to red clover (Trifolium pratense), ryegrass (Lolium perenne), and mustard (Sinapsis alba) [J]. Chemosphere, 2003, 53(8): 993-1003. doi: 10.1016/S0045-6535(03)00584-8 [33] BALDYGA B, WIECZOREK J, SMOCZYNSKI S, et al. Pea plant response to anthracene present in soil [J]. Polish Journal of Environmental Studies, 2005, 14(4): 397-401. [34] HENNER P, SCHIAVON M, DRUELLE V, et al. Phytotoxicity of ancient gaswork soils. Effect of polycyclic aromatic hydrocarbons (PAHs) on plant germination [J]. Organic Geochemistry, 1999, 30(8): 963-969. doi: 10.1016/S0146-6380(99)00080-7 [35] ALKIO M, TABUCHI T M, WANG X C, et al. Stress responses to polycyclic aromatic hydrocarbons in Arabidopsis include growth inhibition and hypersensitive response-like symptoms [J]. Journal of Experimental Botany, 2005, 56(421): 2983-2994. doi: 10.1093/jxb/eri295 [36] 周正宇. 甲基取代多环芳烃的致癌活性与其分子轨道之间的关系 [J]. 曲阜师范大学学报(自然科学版), 1986, 12(2): 56-60. ZHOU Z Y. Relation between carcinogenicty of methylated polycyclic aromatic hydrocarbons and its molecular qrbit [J]. Journal of Qufu Normal University (Natural Science), 1986, 12(2): 56-60(in Chinese).
[37] JERINA D M, LEHR R E. The bay-region theory: a quantum mechanical approach to aromatic hydrocarbon-induced carcinogenicity[M]//Microsomes and Drug Oxidations. Amsterdam: Elsevier, 1977: 709-720. [38] POULSEN M T, LOEW G H. Quantum chemical studies of methyl and fluoro analogs of chrysene: Metabolic activation and correlation with carcinogenic activity [J]. Cancer Biochemistry Biophysics, 1981, 5(2): 81-90. [39] MU J L, JIN F, WANG J Y, et al. The effects of CYP1A inhibition on alkyl-phenanthrene metabolism and embryotoxicity in marine medaka (Oryzias melastigma) [J]. Environmental Science and Pollution Research, 2016, 23(11): 11289-11297. doi: 10.1007/s11356-016-6098-2 [40] HUANG M, ZHANG L, MESAROS C, et al. Metabolism of an alkylated polycyclic aromatic hydrocarbon 5-methylchrysene in human hepatoma (HepG2) cells [J]. Chemical Research in Toxicology, 2015, 28(10): 2045-2058. doi: 10.1021/acs.chemrestox.5b00256 [41] CERNIGLIA C E, LAMBERT K J, MILLER D W, et al. Transformation of 1-and 2-methylnaphthalene by Cunninghamella elegans [J]. Applied and Environmental Microbiology, 1984, 47(1): 111-118. doi: 10.1128/aem.47.1.111-118.1984 [42] VAZQUEZ-DUHALT R, WESTLAKE D W S, FEDORAK P M. Lignin peroxidase oxidation of aromatic compounds in systems containing organic solvents [J]. Applied and Environmental Microbiology, 1994, 60(2): 459-466. doi: 10.1128/aem.60.2.459-466.1994 [43] ARANDA E, ULLRICH R, HOFRICHTER M. Conversion of polycyclic aromatic hydrocarbons, methyl naphthalenes and dibenzofuran by two fungal peroxygenases [J]. Biodegradation, 2010, 21(2): 267-281. doi: 10.1007/s10532-009-9299-2 [44] LUO L J, XIAO Z Y, ZHOU X Y, et al. Quantum chemical calculation to elucidate the biodegradation pathway of methylphenanthrene by green microalgae [J]. Water Research, 2020, 173: 115598. doi: 10.1016/j.watres.2020.115598 [45] 杜兰. 新鞘氨醇杆菌US6-1对溶解态菲及甲基菲生物降解过程的研究[D]. 厦门: 厦门大学, 2017. DU L. Study on the biodegradation of dissolved phenanthrene and methyl phenanthrene by Novosphingobium pentaromativorans US6-1[D]. Xiamen: Xiamen University, 2017(in Chinese).
[46] SIDDIQI M A, YUAN Z X, HONEY S A, et al. Metabolism of PAHs and methyl-substituted PAHs by Sphingomonas paucimobilis strain EPA 505 [J]. Polycyclic Aromatic Compounds, 2002, 22(3/4): 621-630. [47] BUGG T, FOGHT J M, PICKARD M A, et al. Uptake and active efflux of polycyclic aromatic hydrocarbons by Pseudomonas fluorescens LP6a [J]. Applied and Environmental Microbiology, 2000, 66(12): 5387-5392. doi: 10.1128/AEM.66.12.5387-5392.2000 [48] POPHRISTIC V, GOODMAN L. Hyperconjugation not steric repulsion leads to the staggered structure of ethane [J]. Nature, 2001, 411(6837): 565-568. doi: 10.1038/35079036 [49] VOLKMAN J K, ALEXANDER R, KAGI R I, et al. Biodegradation of aromatic hydrocarbons in crude oils from the Barrow Sub-basin of Western Australia [J]. Organic Geochemistry, 1984, 6: 619-632. doi: 10.1016/0146-6380(84)90084-6 [50] HUANG H P, BOWLER B F J, OLDENBURG T B P, et al. The effect of biodegradation on polycyclic aromatic hydrocarbons in reservoired oils from the Liaohe basin, NE China [J]. Organic Geochemistry, 2004, 35(11/12): 1619-1634. [51] WILLIAMS P A, CATTERALL F A, MURRAY K. Metabolism of naphthalene, 2-methylnaphthalene, salicylate, and benzoate by Pseudomonas PG: Regulation of tangential pathways [J]. Journal of Bacteriology, 1975, 124(2): 679-685. doi: 10.1128/jb.124.2.679-685.1975 [52] MAHAJAN M C, PHALE P S, VAIDYANATHAN C S. Evidence for the involvement of multiple pathways in the biodegradation of 1-and 2-methylnaphthalene by Pseudomonas putida CSV86 [J]. Archives of Microbiology, 1994, 161(5): 425-433. doi: 10.1007/BF00288954 [53] SHARANAGOUDA U, KAREGOUDAR T B. Degradation of 2-methylnaphthalene by Pseudomonas sp. strain NGK1 [J]. Current Microbiology, 2001, 43(6): 440-443. doi: 10.1007/s002840010335 [54] DUTTA T K, SELIFONOV S A, GUNSALUS I C. Oxidation of methyl-substituted naphthalenes: Pathways in a versatile Sphingomonas paucimobilis strain [J]. Applied and Environmental Microbiology, 1998, 64(5): 1884-1889. doi: 10.1128/AEM.64.5.1884-1889.1998 [55] NADALIG T, RAYMOND N, NI'MATUZAHROH, et al. Degradation of phenanthrene, methylphenanthrenes and dibenzothiophene by a Sphingomonas strain 2mpII [J]. Applied Microbiology and Biotechnology, 2002, 59(1): 79-85. doi: 10.1007/s00253-002-0960-5 [56] LAMBERTS R F, CHRISTENSEN J H, MAYER P, et al. Isomer-specific biodegradation of methylphenanthrenes by soil bacteria [J]. Environmental Science & Technology, 2008, 42(13): 4790-4796. [57] ZHONG J N, LUO L J, CHEN B W, et al. Degradation pathways of 1-methylphenanthrene in bacterial Sphingobium sp. MP9-4 isolated from petroleum-contaminated soil [J]. Marine Pollution Bulletin, 2017, 114(2): 926-933. doi: 10.1016/j.marpolbul.2016.11.020 [58] DELLAGNEZZE B M, de SOUSA G V, MARTINS L L, et al. Bioremediation potential of microorganisms derived from petroleum reservoirs [J]. Marine Pollution Bulletin, 2014, 89(1/2): 191-200. [59] SEO J S, KEUM Y S, LI Q X. Bacterial degradation of aromatic compounds [J]. International Journal of Environmental Research and Public Health, 2009, 6(1): 278-309. doi: 10.3390/ijerph6010278 [60] TAKIZAWA N, IIDA T, SAWADA T, et al. Nucleotide sequences and characterization of genes encoding naphthalene upper pathway of Pseudomonas aeruginosa PaK1 and Pseudomonas putida OUS82 [J]. Journal of Bioscience and Bioengineering, 1999, 87(6): 721-731. doi: 10.1016/S1389-1723(99)80144-3 [61] DAVIES J I, EVANS W C. Oxidative metabolism of naphthalene by soil pseudomonads. The ring-fission mechanism [J]. The Biochemical Journal, 1964, 91(2): 251-261. doi: 10.1042/bj0910251 [62] TISSOT B P, WELTE D H. Petroleum formation and occurrence[M]. Berlin, Heidelberg: Springer Berlin Heidelberg, 1984. [63] KASAI Y, KISHIRA H, HARAYAMA S. Bacteria belonging to the genus Cycloclasticus play a primary role in the degradation of aromatic hydrocarbons released in a marine environment [J]. Applied and Environmental Microbiology, 2002, 68(11): 5625-5633. doi: 10.1128/AEM.68.11.5625-5633.2002 [64] MUELLER-SPITZ S R, CRAWFORD K D. Silver nanoparticle inhibition of polycyclic aromatic hydrocarbons degradation by Mycobacterium species RJGII-135 [J]. Letters in Applied Microbiology, 2014, 58(4): 330-337. doi: 10.1111/lam.12205 [65] HEDLUND B P, GEISELBRECHT A D, BAIR T J, et al. Polycyclic Aromatic Hydrocarbon Degradation by a New Marine Bacterium, Neptunomonas naphthovorans gen. nov., sp. nov [J]. Applied and Environmental Microbiology, 1999, 65(1): 251-259. doi: 10.1128/AEM.65.1.251-259.1999 [66] OGUNBAYO A, OLANIPEKUN O, OWOADE A. Biodegradation of certain polycyclic hydrocarbons with paenbacillus alvei and penicillum restricum [J]. Journal of Ecological Engineering, 2018, 19(2): 140-148. doi: 10.12911/22998993/81808 [67] 向廷生, 马飞, 潘科. 轻中度生物降解作用对原油中烷基萘和烷基菲的影响 [J]. 西安石油大学学报(自然科学版), 2012, 27(1): 81-86,122. XIANG T S, MA F, PAN K. Effect of mild-to-moderate biodegradation on alkyl naphthalene and alkyl phenanthrene in crude oil [J]. Journal of Xi'an Shiyou University (Natural Science Edition), 2012, 27(1): 81-86,122(in Chinese).
[68] MUSAT F, GALUSHKO A, JACOB J, et al. Anaerobic degradation of naphthalene and 2-methylnaphthalene by strains of marine sulfate-reducing bacteria [J]. Environmental Microbiology, 2009, 11(1): 209-219. doi: 10.1111/j.1462-2920.2008.01756.x [69] BERDUGO-CLAVIJO C, DONG X L, SOH J, et al. Methanogenic biodegradation of two-ringed polycyclic aromatic hydrocarbons [J]. FEMS Microbiology Ecology, 2012, 81(1): 124-133. doi: 10.1111/j.1574-6941.2012.01328.x [70] FOLWELL B D, MCGENITY T J, PRICE A, et al. Exploring the capacity for anaerobic biodegradation of polycyclic aromatic hydrocarbons and naphthenic acids by microbes from oil-sands-process-affected waters [J]. International Biodeterioration & Biodegradation, 2016, 108: 214-221. [71] BUDZINSKI H, GARRIGUES P, CONNAN J, et al. Alkylated phenanthrene distributions as maturity and origin indicators in crude oils and rock extracts [J]. Geochimica et Cosmochimica Acta, 1995, 59(10): 2043-2056. doi: 10.1016/0016-7037(95)00125-5 [72] RADKE M. Application of aromatic compounds as maturity indicators in source rocks and crude oils [J]. Marine and Petroleum Geology, 1988, 5(3): 224-236. doi: 10.1016/0264-8172(88)90003-7 [73] SAMANTA S K, SINGH O V, JAIN R K. Polycyclic aromatic hydrocarbons: Environmental pollution and bioremediation [J]. Trends in Biotechnology, 2002, 20(6): 243-248. doi: 10.1016/S0167-7799(02)01943-1 [74] GILEWICZ M, NADALIG T, BUDZINSKI H, et al. Isolation and characterization of a marine bacterium capable of utilizing 2-methylphenanthrene [J]. Applied Microbiology and Biotechnology, 1997, 48(4): 528-533. doi: 10.1007/s002530051091 [75] SABATÉ J, GRIFOLL M, VIÑAS M, et al. Isolation and characterization of a 2-methylphenanthrene utilizing bacterium: Identification of ring cleavage metabolites [J]. Applied Microbiology and Biotechnology, 1999, 52(5): 704-712. doi: 10.1007/s002530051582 [76] SHA S, ZHONG J N, CHEN B W, et al. Novosphingobium guangzhouense sp. nov., with the ability to degrade 1-methylphenanthrene [J]. International Journal of Systematic and Evolutionary Microbiology, 2017, 67(2): 489-497. doi: 10.1099/ijsem.0.001669 [77] WANG X W, CAI T, WEN W T, et al. Surfactin for enhanced removal of aromatic hydrocarbons during biodegradation of crude oil [J]. Fuel, 2020, 267: 117272. doi: 10.1016/j.fuel.2020.117272 [78] NADALIG T, RAYMOND N, GILEWICZ M, et al. Development of a protocol to study aerobic bacterial degradation of polycyclic aromatic hydrocarbons: Application to phenanthrenes [J]. Polycyclic Aromatic Compounds, 2000, 18(2): 177-192. doi: 10.1080/10406630008028144 [79] ROWLAND S J, ALEXANDER R, KAGI R I, et al. Microbial degradation of aromatic components of crude oils: A comparison of laboratory and field observations [J]. Organic Geochemistry, 1986, 9(4): 153-161. doi: 10.1016/0146-6380(86)90065-3 [80] BUDZINSKI H, NADALIG T, RAYMOND N, et al. Evidence of two metabolic pathways for degradation of 2-methylphenanthrene by Sphingomonas sp. strain (2mpii) [J]. Environmental Toxicology and Chemistry, 2000, 19(11): 2672. doi: 10.1002/etc.5620191109 [81] KELLEY I, FREEMAN J P, CERNIGLIA C E. Identification of metabolites from degradation of naphthalene by a Mycobacterium sp [J]. Biodegradation, 1990, 1(4): 283-290. doi: 10.1007/BF00119765 [82] 曹晓星, 田蕴, 胡忠, 等. PAHs降解基因及降解酶研究进展 [J]. 生态学杂志, 2007, 26(6): 917-924. doi: 10.3321/j.issn:1000-4890.2007.06.026 CAO X X, TIAN Y, HU Z, et al. Research progress in PAHs degradation genes and enzymes [J]. Chinese Journal of Ecology, 2007, 26(6): 917-924(in Chinese). doi: 10.3321/j.issn:1000-4890.2007.06.026
[83] BUTLER C S, MASON J R. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases [J]. Advances in Microbial Physiology, 1996, 38: 47-84. [84] JIANG H, PARALES R E, LYNCH N A, et al. Site-directed mutagenesis of conserved amino acids in the alpha subunit of toluene dioxygenase: Potential mononuclear non-heme iron coordination sites [J]. Journal of Bacteriology, 1996, 178(11): 3133-3139. doi: 10.1128/jb.178.11.3133-3139.1996 [85] LANGE S J, QUE L Jr. Oxygen activating nonheme iron enzymes [J]. Current Opinion in Chemical Biology, 1998, 2(2): 159-172. doi: 10.1016/S1367-5931(98)80057-4 [86] SANAKIS Y, MAMMA D, CHRISTAKOPOULOS P, et al. Catechol 1, 2-dioxygenase from Pseudomonas putida in organic media—an electron paramagnetic resonance study [J]. International Journal of Biological Macromolecules, 2003, 33(1/2/3): 101-106. [87] TØNDERVIK A, BRUHEIM P, BERG L, et al. Ralstonia sp U2 naphthalene dioxygenase and Comamonas sp JS765 nitrobenzene dioxygenase show differences in activity towards methylated naphthalenes [J]. Journal of Bioscience and Bioengineering, 2012, 113(2): 173-178. doi: 10.1016/j.jbiosc.2011.10.001 [88] FANG T T, PAN R S, JIANG J, et al. Effect of salinity on community structure and naphthalene dioxygenase gene diversity of a halophilic bacterial consortium [J]. Frontiers of Environmental Science & Engineering, 2016, 10(6): 1-8. [89] BEN SAID O, GOÑI-URRIZA M S, EL BOUR M, et al. Characterization of aerobic polycyclic aromatic hydrocarbon-degrading bacteria from Bizerte lagoon sediments, Tunisia [J]. Journal of Applied Microbiology, 2008, 104(4): 987-997. doi: 10.1111/j.1365-2672.2007.03621.x [90] OKUTA A, OHNISHI K, YAGAME S, et al. Intersubunit interaction and catalytic activity of catechol 2, 3-dioxygenases[J]. The Biochemical Journal, 2003, 371(Pt 2): 557-564. [91] CRUTCHER S E, GEARY P J. Properties of the iron–sulphur proteins of the benzene dioxygenase system from Pseudomonas putida [J]. Biochemical Journal, 1979, 177(2): 393-400. doi: 10.1042/bj1770393 [92] YEH W K, GIBSON D T, LIU T N. Toluene dioxygenase: A multicomponent enzyme system [J]. Biochemical and Biophysical Research Communications, 1977, 78(1): 401-410. doi: 10.1016/0006-291X(77)91268-2 [93] 张丹, 李兆格, 包新光, 等. 细菌降解萘、菲的代谢途径及相关基因的研究进展 [J]. 生物工程学报, 2010, 26(6): 726-734. doi: 10.13345/j.cjb.2010.06.011 ZHANG D, LI Z G, BAO X G, et al. Recent advances in bacterial biodegradation of naphthalene, phenanthrene by bacteria: A review [J]. Chinese Journal of Biotechnology, 2010, 26(6): 726-734(in Chinese). doi: 10.13345/j.cjb.2010.06.011
[94] HABE H, OMORI T. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria [J]. Bioscience, Biotechnology, and Biochemistry, 2003, 67(2): 225-243. doi: 10.1271/bbb.67.225 [95] HARAYAMA S, REKIK M, WASSERFALLEN A, et al. Evolutionary relationships between catabolic pathways for aromatics: Conservation of gene order and nucleotide sequences of catechol oxidation genes of pWW0 and NAH7 plasmids [J]. Molecular and General Genetics MGG, 1987, 210(2): 241-247. doi: 10.1007/BF00325689 [96] ZYLSTRA G J, KIM E, GOYAL A K. Comparative molecular analysis of genes for polycyclic aromatic hydrocarbon degradation [J]. Genetic Engineering, 1997, 19: 257-269. doi: 10.1007/978-1-4615-5925-2_14 [97] SAITO A, IWABUCHI T, HARAYAMA S. A novel phenanthrene dioxygenase from Nocardioides sp Strain KP7: Expression in Escherichia coli. [J]. Journal of Bacteriology, 2000, 182(8): 2134-2141. doi: 10.1128/JB.182.8.2134-2141.2000 [98] LAURIE A D, LLOYD-JONES G. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism [J]. Journal of Bacteriology, 1999, 181(2): 531-540. doi: 10.1128/JB.181.2.531-540.1999 [99] 张维荣, 严康, 汪海珍, 等. 基于1983—2019年文献计量对多环芳烃降解基因研究及进展的剖析 [J]. 环境科学学报, 2020, 40(3): 1138-1148. doi: 10.13671/j.hjkxxb.2019.0359 ZHANG W R, YAN K, WANG H Z, et al. Bibliometric analysis of research progress on polycyclic aromatic hydrocarbons-degrading genes during 1983—2019 [J]. Acta Scientiae Circumstantiae, 2020, 40(3): 1138-1148(in Chinese). doi: 10.13671/j.hjkxxb.2019.0359
[100] KULAKOV L A, CHEN S C, ALLEN C C R, et al. Web-type evolution of Rhodococcus gene clusters associated with utilization of naphthalene [J]. Applied and Environmental Microbiology, 2005, 71(4): 1754-1764. doi: 10.1128/AEM.71.4.1754-1764.2005 [101] LARKIN M J, KULAKOV L A, ALLEN C C. Biodegradation and Rhodococcus - Masters of catabolic versatility [J]. Current Opinion in Biotechnology, 2005, 16(3): 282-290. doi: 10.1016/j.copbio.2005.04.007 [102] KUMARI S, REGAR R K, BAJAJ A, et al. Simultaneous biodegradation of polyaromatic hydrocarbons by a Stenotrophomonas sp: Characterization of nid genes and effect of surfactants on degradation [J]. Indian Journal of Microbiology, 2017, 57(1): 60-67. doi: 10.1007/s12088-016-0612-6 [103] KIM S J, KWEON O, JONES R C, et al. Complete and integrated Pyrene degradation pathway in Mycobacterium vanbaalenii PYR-1 based on systems biology [J]. Journal of Bacteriology, 2007, 189(2): 464-472. doi: 10.1128/JB.01310-06 [104] PAGNOUT C, FRACHE G, POUPIN P, et al. Isolation and characterization of a gene cluster involved in PAH degradation in Mycobacterium sp. strain SNP11: Expression in Mycobacterium smegmatis mc2155 [J]. Research in Microbiology, 2007, 158(2): 175-186. doi: 10.1016/j.resmic.2006.11.002 [105] KRIVOBOK S, KUONY S, MEYER C, et al. Identification of Pyrene-induced proteins in Mycobacterium sp strain 6PY1: Evidence for two ring-hydroxylating dioxygenases [J]. Journal of Bacteriology, 2003, 185(13): 3828-3841. doi: 10.1128/JB.185.13.3828-3841.2003 [106] WU M Y, KWOK Y H, ZHANG Y G, et al. Synergetic effect of vacuum ultraviolet photolysis and ozone catalytic oxidation for toluene degradation over MnO2-rGO composite catalyst [J]. Chemical Engineering Science, 2021, 231: 116288. doi: 10.1016/j.ces.2020.116288 [107] BURLAGE R S, HOOPER S W, SAYLER G S. The TOL (pWW0) catabolic plasmid [J]. Applied and Environmental Microbiology, 1989, 55(6): 1323-1328. doi: 10.1128/aem.55.6.1323-1328.1989 [108] NIKEL P I, SILVA-ROCHA R, BENEDETTI I, et al. The private life of environmental bacteria: Pollutant biodegradation at the single cell level [J]. Environmental Microbiology, 2014, 16(3): 628-642. doi: 10.1111/1462-2920.12360 [109] WOLFE M D, ALTIER D J, STUBNA A, et al. Benzoate 1, 2-dioxygenase from Pseudomonas putida: single turnover kinetics and regulation of a two-component rieske dioxygenase [J]. Biochemistry, 2002, 41(30): 9611-9626. doi: 10.1021/bi025912n [110] NEIDLE E, HARTNETT C, ORNSTON L N, et al. Cis-diol dehydrogenases encoded by the TOL pWW0 plasmid xylL gene and the Acinetobacter calcoaceticus chromosomal benD gene are members of the short-chain alcohol dehydrogenase superfamily [J]. European Journal of Biochemistry, 1992, 204(1): 113-120. doi: 10.1111/j.1432-1033.1992.tb16612.x [111] EATON R W. P-Cymene catabolic pathway in Pseudomonas putida F1: Cloning and characterization of DNA encoding conversion of p-cymene to p-cumate [J]. Journal of Bacteriology, 1997, 179(10): 3171-3180. doi: 10.1128/jb.179.10.3171-3180.1997 [112] SUHARA K, TAKEMORI S, KATAGIRI M. The purification and properties of benzylalcohol dehydrogenase from Pseudomonas SP [J]. Archives of Biochemistry and Biophysics, 1969, 130: 422-429. doi: 10.1016/0003-9861(69)90054-X [113] MECKENSTOCK R U, SAFINOWSKI M, GRIEBLER C. Anaerobic degradation of polycyclic aromatic hydrocarbons [J]. FEMS Microbiology Ecology, 2004, 49(1): 27-36. doi: 10.1016/j.femsec.2004.02.019 [114] PÉREZ-JIMÉNEZ J R, YOUNG L Y, KERKHOF L J. Molecular characterization of sulfate-reducing bacteria in anaerobic hydrocarbon-degrading consortia and pure cultures using the dissimilatory sulfite reductase (dsrAB) genes [J]. FEMS Microbiology Ecology, 2001, 35(2): 145-150. doi: 10.1111/j.1574-6941.2001.tb00798.x -



 下载:
下载: