-
电化学厌氧消化(electrochemical anaerobic digestion,EAD)是一种利用微生物电解池(microbial electrolysis cell,MEC)的耦合厌氧消化(anaerobic digestion,AD)的新技术,其原理是利用MEC的制氢功能,对系统内的二氧化碳(CO2)进行固定和转化,生成更多的CH4,从而减少CO2的排放[1],并极大地提高了CH4的转化率和沼气的品质[2],所以,EAD作为一种新型的沼气制备技术,因其具有甲烷转化率高、能量回收率高、能耗低、适用于多种有机废弃物和废水的降解等优势而受到了广泛的关注,是目前AD领域的最新发现[3-5].
但不管是AD,还是EAD,绝大多数相关研究仍然停留在提高消化能力或者副产物的产率等方面[6-8],很少涉及代谢过程的研究. 代谢通量分析(metabolic flux analysis,MFA)是一项研究胞内代谢状态和分析系统代谢能力的强有力技术,通过MFA:(1)可以确定细胞或者系统内存在的不同代谢途径,明确物质的流向和流量[8];(2)可以计算胞内的中间产物的通量,对未知途径加以辨别[9];(3)可以识别代谢节点的刚柔性[10],和对环境扰动的影响做出评价,从而对代谢途径中具有特殊地位的途径实施有效调控[11](如优化目标产物代谢途径、计算最大理论转化率等[12]). 因此,本文将利用MFA中的通过通量平衡分析(flux balance analysis,FBA)的方法,构建EAD代谢网络,研究EAD代谢网络中的相关代谢途径的通量分布信息.
在代谢通量分析中,通量是反映各代谢产物在整个代谢网络中参与程度的重要参数[7],可以依据通量的大小评价各代谢途径在整个代谢网络中所发挥的作用[13]. 然而由于EAD代谢网络中的相关代谢途径,往往是在相应的酶催化下进行的,在EAD代谢途径中,乙酸的形成和转化是EAD产甲烷过程中最为关键的节点. 乙酸的形成主要的途径是乙酰辅酶A通过磷酸转乙酰酶(PTA)转化为乙酰基-P,然后再通过乙酸激酶(AK)从乙酰基-P转化为乙酰基-P,AK催化反应产生ATP[14]. 厌氧消化EMP途径所产生的丙酮酸转化为乙醛和二氧化碳,然后乙醛通过ADH还原成乙醇[15]. 丁酸的产生则与磷酸转丁酰酶(PTB)与丁酸激酶相关. 此外,ADP/ATP转运酶也发挥着重要的作用. ADP/ATP转运酶又被称为腺嘌呤核苷酸易位子或ADP/ATP载体蛋白,ADP/ATP转运酶通过ADP 与ATP相互转化产生能量从而为细胞提供能量[16]. 而在乙酸转化中,CoF420参与产甲烷菌中的氧化还原反应,充当电子载体. CoF430是甲基辅酶M还原酶的辅助因子,在甲烷生成的最后一步释放甲烷. 而辅酶M是古菌产甲烷菌代谢中甲基转移反应所需的辅酶.
综上所述,利用FBA不仅可以对EAD系统的代谢过程进行表征,还能对代谢通量进行分析和直观地了解代谢网络中不同代谢途径的产物生成情况以及代谢关键节点处的酶活性变化水平.
-
EAD实验装置由 电解电流及电压记录单元、 EAD反应单元和排水集气单元组成,EAD反应单元主要包含400 mL的发酵罐,其中电极距离为3.0 cm,一端的电极片完全地浸于发酵液中,另一端通过导线与宽屏无纸记录仪(SIN-R7000A,中国)相连;在EAD系统中,阳极为石墨电极,阴极为铂电极,参比电极为饱和的Ag/AgCl电极. EAD实验装置如图1所示.
-
厌氧活性污泥来自云南师范大学昆明实验室通过猪粪厌氧发酵. 经检测,pH值为8.07,TS为17.11%,VS为10.08%.
缓冲营养液配比:NaH2PO4∙2H2O(5.54 g·L−1)、Na2HPO4∙12H2O(23.09 g·L−1)、KCl(0.26 g·L−1)和NH4Cl(0.62 g·L−1).
微量元素液配比:Na2WO4∙2H2O(0.03 g·L−1)、NaCl(1 g·L−1)、FeCl2∙7H2O(0.07 g·L−1)、CuSO4∙5H2O(0.01 g·L−1)、NiCl2∙6H2O(0.02 g·L−1)、C6H9NO6(1.50 g·L−1)、MgSO4(3.00 g·L−1)、AlK(SO4)2∙12H2O(0.01 g·L−1)、ZnCl2(0.13 g·L−1)、CaCl2∙2H2O(0.10 g·L−1)、H3BO3(0.01 g·L−1)、MnCl2∙4H2O(0.60 g·L−1)、CoCl2∙6H2O(0.10 g·L−1)、Na2MoO4(0.03 g·L−1)和复合维生素(1 粒·L−1).
-
EAD实验设置3个实验组,每组3个平行. 首先在每个实验组的发酵罐中添加:葡萄糖1.0 g,接种物120.0 g,缓冲营养液100.0 mL,微量元素液10.0 mL,再补加蒸馏水使物料的总质量为360.0 g;其次将密封好的发酵罐连接气体收集装置,再利用高纯氮排出体系内的溶解氧和空气(通气时间为3 min),使EAD体系处于厌氧状态;最后施加恒定电压1.0 V,进行培养.
培养阶段:将发酵罐置于恒温磁力搅拌器中,设置温度为30.0 ℃;每天监测pH值变化,待其恢复到7.0左右,立即添加等量的葡萄糖将体系培养至稳定.
扰动阶段:培养阶段结束后,测定发酵液中底物和代谢产物的浓度,同时记下该时间点为扰动的初始点,随后添加1.0 g的葡萄糖,施加扰动后,通过监测体系中底物的消耗量和代谢产物的生成量,然后进行代谢通量分析.
-
日产气量:采用排水法法,每天定时记录产气量. TS和VS:在(105±5)℃下干燥并在(550±10)℃下马弗炉燃烧1 h后,测量污泥的. pH值:采用pH计(HAOSHI H-1008A,中国).
VFAs:样品经离心分离,取上清液,采用气相色谱仪(FULI, GC9790II,中国),以FID为检测器进行测量[17]. 色谱柱为KB- FFAP(30 m×0. 32 mm×0. 25 μm)毛细管柱.
气体含量:采用气相色谱仪(FULI, GC9790II,中国),以TCD 检测器对H2、CH4和CO2含量进行测量. 色谱柱为TDX-01填充柱. 葡萄糖含量:采用3,5-二硝基水杨酸比色法测定[18].
-
酶活测量采用改良的双抗体夹心酶联免疫吸附测定(ELISA)方法[19-21]在测量过程中,以ADP/ATP转运酶、磷酸转乙酰酶(PTA)、乙酸激酶(AK)、磷酸转丁酰酶(PTB)、丁酸激酶(BK)、辅酶M(CoM)、甲基丙二酰CoA变位酶(MCM)、辅酶F420(F420)、辅酶F430(F430)为标准酶. 检测时在酶标包被板上设定空白孔、标准孔、待测样品孔. 空白孔不加样品及标准酶,作为对照,标准孔加标准酶,待测样品孔中先加酶试剂盒中的样品稀释液,然后再加待测样品. 按照酶试剂盒说明书进行操作,最后采用Epoch酶标仪在450 nm波长下进行测定,以空白孔调零依序测量各孔的吸光度(OD值),并通过标准曲线计算样品中酶的活性.
-
通量分析采用通量平衡分析(FBA)的方法,采用代谢通量分析软件Cell Net Analyzer进行代谢网络的构建及分析. FBA是MFA中基于质量守恒的代谢过程研究常用的一种数学建模分析法[22],在拟稳态条件下,模型中代谢产物处于平衡状态,此时,可将代谢物的通量描述为一个线性方程,只需对底物和相关产物的浓度变化进行测量,再采用线性规划或者计量矩阵分析方法则可确定每种产物的代谢通量[23].
-
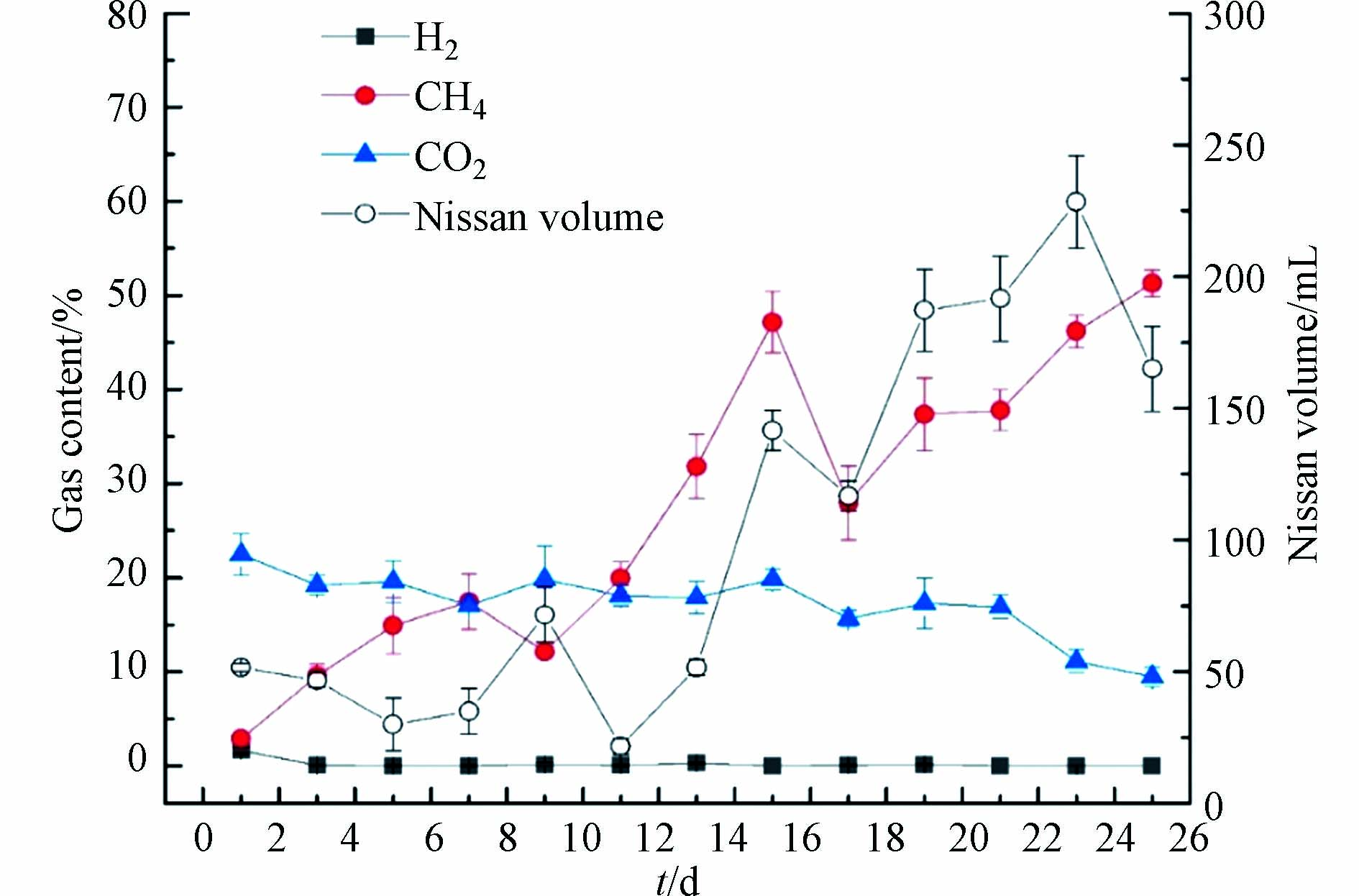
EAD反应过程中产气特性的变化是表征反应器效能的重要因素. 图2为培养过程中的产气变化情况,从图2可以发现,在反应器启动阶段,气体中以CO2为主,随着反应器的运行,产甲烷菌逐渐生长富集. 在稳定阶段,其甲烷含量最高可以达到55%左右,葡萄糖厌氧消化中理论产甲烷含量在50%左右,也就是说,MEC的引入明显提升了葡萄糖的理论产甲烷率,而且此时CO2的含量降低到11%左右,明显的降低了CO2的排放. 在反应器稳定后,日产气量也得到了明显的提升,从最初的50 mL·d−1左右增加到了200 mL·d−1左右. 对于氢气含量,除启动阶段有氢气产生外,反应中后期基本没有氢气产生,这可能是由于前期pH较低,适于产氢菌的生长,中后期pH值恢复较快,可以迅速恢复至已6.5以上,不利于产氢菌的生长,导致氢气含量极低. 此外,也可能是由于电化学辅助促进了H2还原CO2途径产甲烷,使得反应器中的H2迅速被噬氢产甲烷菌消耗,从而提高了CH4的含量,降低了H2的含量.
-
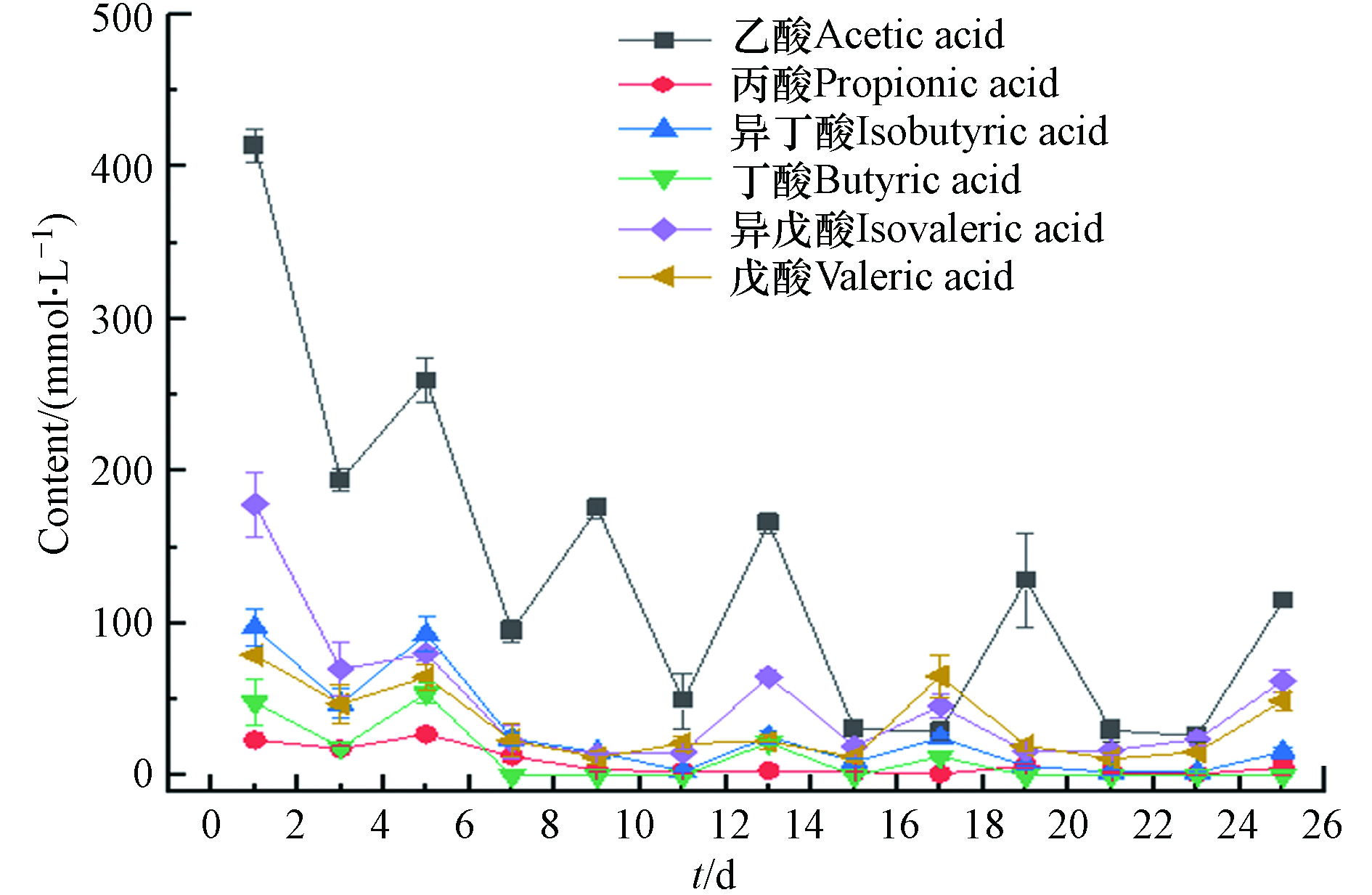
VFAs是厌氧消化中重要的中间产物,其产生和分解过程受多种因素影响,同时也关系到EAD产甲烷过程的稳定性. EAD体系中 VFAs的浓度一旦超过 200 mmol·L−1 就有可能发生酸抑制问题[24],并对产甲烷过程产生严重的影响,然而本次实验研究并未出现VFAs累积的现象. 图3显示了EAD培养过程中各短链有机酸的变化情况. 从图3可以看出,各短链有机酸的变化与底物补充时间相关,培养前期VFAs波动较大且浓度较高,但15 d后,因阳极上形成了电活性微生物膜,致使VFAs能够在阳极表面降解,尽管进料时间间隔缩短为3 d,也没有产生VFAs累积,表明EAD具有较强的抗酸化的能力.
在厌氧消化中丁酸累积是抑制厌氧消化的重要因素之一,由于丁酸的产氢产乙酸过程必须依赖氢还原CO2产甲烷过程,否则本身不能自发进行,一旦产氢产乙酸和产甲烷过程失衡,将会产生严重的丁酸抑制作用。然而在EAD中丁酸的降解除了依赖氢还原CO2产甲烷过程外,还可以在电活性微生物的作用下,在电极表面降解. 在这条途径中,丁酸分解为CO2、H+和电子, H+向阴极迁移在阴极上获得电子而形成H2,这部分H2同样被氢营养型产甲烷菌利用还原CO2,加强H2还原CO2产甲烷途径.
-
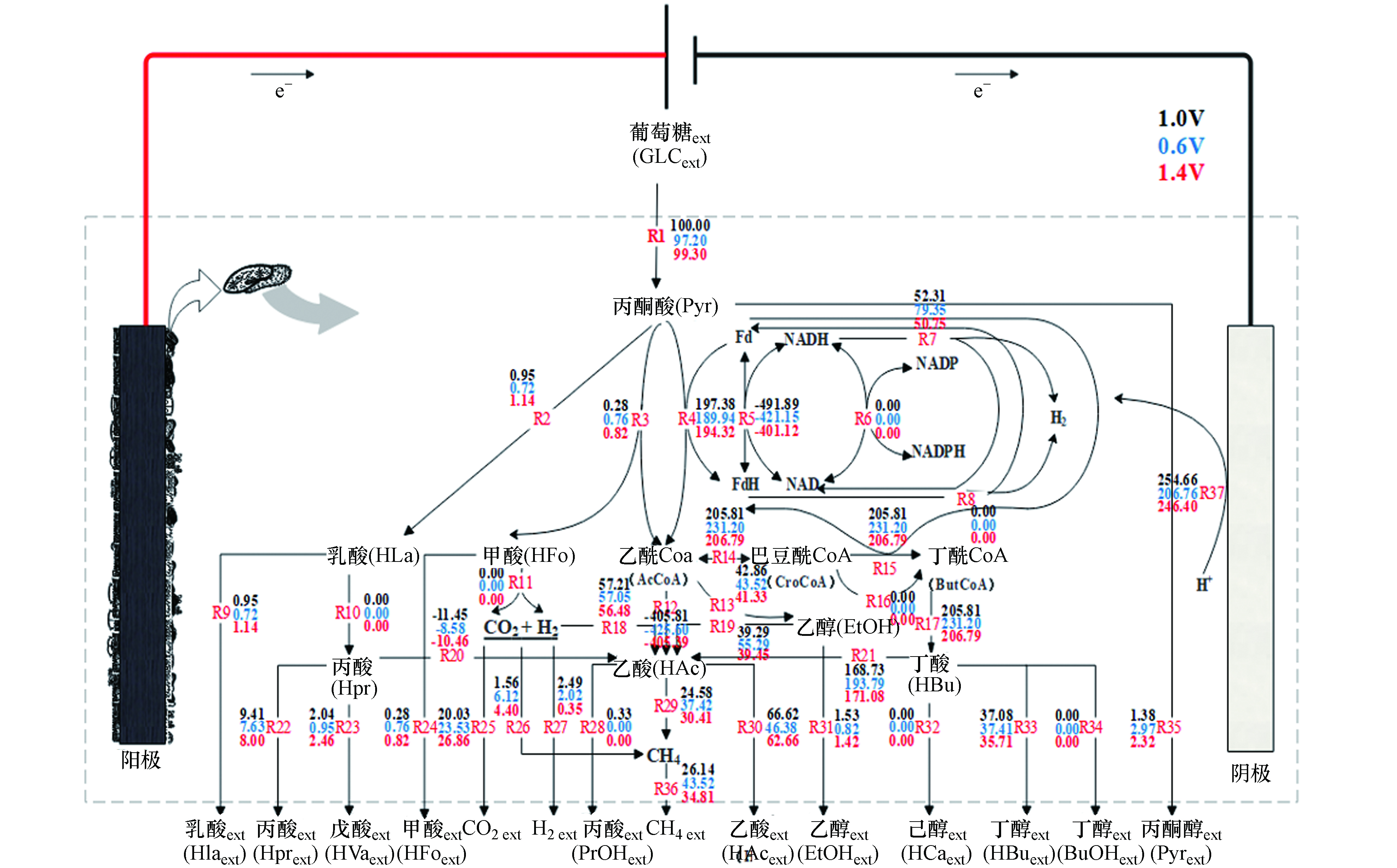
对于纯培养体系而言,由于体系内菌种单一,代谢产物和途径比较明确,构建与之相对应的代谢网络相对比较容易. 但对于复杂的混合培养而言,特别是像EAD这种由多种微生物构成的体系,变量较多,对于代谢网络的构建及分析难度较大[25],为了使代谢通量分析能适用于混合体系,提出了“统一微生物”这一个概念[26],同时国内外一些学者已经在部分混合体系中作了应用上的尝试,并且成功地建立了发酵制氢体系的代谢网络[27],还有在新型多级厌氧甲烷反应器中也进行了FBA的研究[28],但还需进一步地完善,才能使其适用于EAD的混合培养体系中. EAD是在AD中引入MEC,由于MEC的引入,使H+向阴极迁移,在阴极上生成H2,这在AD基础上增加了一条重要的产氢途径. 根据相关研究表明[9, 12],在严格的厌氧条件下,丙酮酸几乎不会流向三羧酸循环,甲酸也不是所有严格厌氧菌都能产生. 结合相关研究[29-33],构建了如图4所示的EAD代谢网络该网络包含了37个主要的代谢反应式,每一步的代谢的具体生化反应过程如表1所示.
-
在培养阶段中,当EAD运行稳定后,从取样口取样,检测残余葡萄糖和相关代谢物的含量,然后添加1 g的葡萄糖,以此时的状态为FBA的初始状态,实施电压扰动. 在对照组电压为1.0 V的基础上,扰动组1电压降低至0.6 V,而扰动组2增加至1.4 V. 14 h后,再次检测残余葡萄糖和相关代谢物,产气量和气体含量. 经过计算得到EAD扰动前后的发酵液中代谢产物的净生成速率,如表2所示.
根据表2中的净生成速率,采用运行Cell Net Analyzer软件,获得EAD代谢通量分布信息,如图4所示,并以H2、CO2、HAc和CH4代谢物进行评价.
在0.6 V扰动下,NADH通量明显优于1.4 V电压扰动,且此条件下最终产物甲烷通量达到43.52,明显高于1.0 V和1.4 V下的26.14和34.81. 由图4可知,H2的代谢途径主要形成途径为:R7、R20、R21和R37. 由通量值可知,EAD中H2主要来源于电极反应R37,和丁酸降解为乙酸的途径R21,其次是NADH平衡调节产氢R7. CH4为最终的目标代谢物,主要由H2还原CO2途径R26和乙酸转化途径R29两条途径形成. EAD体系中CH4的合成途径是以乙酸转化途径占主导地位,其次为H2还原CO2途径,1.0 V下EAD体系的这两条途径的占CH4产生总量的64.95%和35.05%. 0.6 V和1.4 V扰动下,通过H2还原CO2产生的产甲烷(R26)分别占13.44%和12.76%,相比对照组5.97%均有所提高,表明电压扰动使得EAD产甲烷代谢途径偏向了氢营养型产甲烷. 值得注意的EAD产甲烷系统明显产生了甲酸,但根据代谢通量,CH4和H2并无一来源于甲酸,说明EAD反应体系中可能没有噬甲酸型产甲烷菌存在. 最终产物H2大多由EAD中的阴极产生,但在最终产物中,氢气通量明显下降,说明H2还原为CH4(R26)的途径较为活跃. 本次从发酵液代谢物中未检测到己酸、丙醇和己酸盐. 乙酸的来源有R12、R18、R19、R20和R21途径. 通量值表明乙酰辅酶A转化是乙酸最主要的来源,其次是丁酸转化、H2还原CO2、乙醇转化和丙酸转化. 其通量大小受多个中间产物的影响,但在电压扰动(0.6 V和1.4 V)下,其通量明显增大,其中以0.6 V扰动增幅最大,说明反应器中的产酸微生物在此时相对较活跃. 乳酸和乙醇的产生不利于H2的生产,图中0.6 V扰动下的乳酸通量较高,这也可能是导致了H2较低的原因之一.
-
表3为葡萄糖代谢过程中关键酶活性的变化. 其中ADP/ATP转运酶的活性,0.6 V和1.4 V扰动时的酶活性均高于对照组1.0 V 的239.28 IU·L−1. ADP/ATP是能量代谢的载体,为微生物生长和繁殖提供能量基础. 当电压扰动时,需要大量能量来使微生物群落发生转变,因此ADP/ATP转运酶酶活性增加. 在乙酸代谢途径中首要的是乙酸的活化,它需要由乙酸激酶AK和磷酸转乙酰酶PTA的先后催化来进行. 在ATP参与下由乙酸激酶AK催化,乙酸被磷酸化为乙酰磷酸,然后乙酰磷酸再在磷酸转乙酰酶PTA催化下生成乙酰辅酶A,进行乙酸代谢[34-35]. 但从图5中可以看出,所产生的乙酸大部分都R12途径逆反应生成乙酰辅酶A,其中当进行0.6 V扰动时,R12逆反应通量最大,这与在0.6 V时PTA活性达到115.69 U·L−1检测结果相吻合. 当进行电压扰动时,EAD体系内在乙酸代谢中PTA和AK的酶活调节现象相似,有研究表明[36]当谷氨酸棒杆菌生长在以乙酸(取代葡萄糖)为碳源的培养基上时,PTA和AK酶活性明显提高,该现象是微生物碳代谢中葡萄糖效应的具体体现,表明酶在基因表达环节上可能存在葡萄糖基质上的负调控效应或乙酸基质上的正调控效应. 降低电压,PTA与AK酶活性增加. PTB和BK酶与丁酸产生有关,由图5代谢通量可知,丁酸的产生是从乙酰基通过PTB与BK的共同催化所形成. 在丁酸代谢中,PTB催化反应产生大量的丁酰辅酶A,但产生的丁酸大部分却通过R21转化为乙酸. 在0.6 V扰动下,PTB的酶活性352.19 U·L−1,明显高于对照组310.19 U·L−1和1.4 V的338.04 U·L−1,但BK酶活性,对照组1.0 V的酶活性52.59 U·L−1,要略高于0.6 V的50.92 U·L−1. PTB酶活性高于BK,这是由于PTB需要先将丁酰辅酶A转化为在丁酰磷酸,最终再由BK催生与ADP反应形成丁酸和ATP. 而BK在1.0 V与0.6 V酶活性差异不大,这是可能是由于电压BK影响不大. 辅酶M在甲烷形成的最终反应步骤中充当甲基的载体. CoM的化学结构虽然很简单,但在甲基还原酶的反应体系中却有高度的专一性,因此在进行电压扰动时,辅酶M的酶活性变化差异不大. 值得一提是,MCM酶活性变化与CoM酶活性变化相似. 在丙酮酸转化为丙酸代谢过程中,关键酶为MCM,在电压扰动下,酶活性变化差异较小. 结合图6中的产丙酸代谢通量图可以看出,丙酸通量变化也差异较小,说明酶活性与所构建的代谢通量有较好的相关性. CoF420在产甲烷途径中电子载体,而CoF430产甲烷古菌形成甲烷最后步骤作为甲基载体的四吡咯化合物,因此产甲烷的总通量应当取决于CoF420与CoF430共同酶活性,在0.6 V电压扰动下,总酶活性为63.01 IU·L−1,对照组为56.94 IU·L−1,1.4 V扰动为59.57 IU·L−1,这与图5中产甲烷代谢通量中,0.6 V时产甲烷最多,1.4 V次之,对照组最少结果相吻合.
-
本研究通过外加电压对EAD体系扰动过程中代谢通量的影响,对EAD代谢网络通量上的变化结合关键酶活性的变化,进一步分析电压扰动对EAD系统代谢产物生成上的提高或者降低提供理论依据,由通量分析结果可知,EAD体系中并没有很高的H2生成,相反,EAD体系中产甲烷能力得到提高. 原因可能是由于电解协助装置的引入,极大地促进H2的产生,随后,所产生的H2被用来还原CO2产乙酸和产甲烷的代谢过程,进而提升了EAD系统的产甲烷能力. 结合关键节点处酶活性变化,酶活性与代谢通量有着较好的相关性. 在进行电压扰动时,ADP/ATP转运酶、PTA与AK、PTB与BK,这些酶活性均随电压变化而产生较大的差异. 而CoM与MCM酶活性,具有高度专一性的酶,随电压扰动变化不大. 在0.6 V电压扰动下,CoF420与CoF430酶活性与产甲烷代谢通量变化相吻合.
电化学厌氧消化代谢途径中关键节点处酶活性的研究
Study on enzyme activity at key nodes in EAD metabolic pathway
-
摘要: 本研究以活性污泥作为接种物,葡萄糖作为底物,电解电压作为扰动因素,采用通量平衡分析(FBA)的方法,构建电化学厌氧消化(EAD)的代谢网络,并对其通量平衡分析和关键代谢节点的信息进行研究. 此外,针对特定关键节点的通量变化情况,结合EAD体系中关键酶活性变化进行分析. 实验结果表明,通过对EAD体系的FBA和关键节点进行研究,发现EAD体系中CH4的合成途径是以乙酸转化途径占主导地位,其次为H2还原CO2途径,EAD体系的这两条途径的占CH4产生总量的64.95%和35.05%. 在关键酶活性检测中,关键酶的活性与产甲烷途径显示出较好的一致性. 在0.6 V扰动下,关键酶活性都有所增加,催化微生物体内反应加快,与代谢通量变化有较高的相关性. 研究还发现在电压扰动下EAD体系中H2还原CO2产生的产甲烷通量13.44%和12.76%均高于对照组5.97%,进而提升了EAD系统的整体产甲烷能力.
-
关键词:
- 电化学厌氧消化(EAD) /
- 代谢通量分析 /
- 酶活性.
Abstract: In this study, activated sludge was used as the inoculum, glucose was used as the substrate, and the electrolytic voltage was used as the disturbance factor. The method of flux balance analysis (FBA) was used to construct the metabolic network of EAD, and its flux balance analysis (FBA) and key Metabolic node information for research. In addition, the flux changes of specific key nodes were analyzed in combination with the changes of key enzyme activities in the EAD system. The experimental results show that: by studying the FBA and key nodes of the EAD system, it is found that the synthesis pathway of CH4 in the EAD system is dominated by the acetate conversion pathway, followed by the H2 reduction CO2 pathway, and the two pathways in the EAD system account for CH4. Generated 64.95% and 35.05% of the total. In the detection of key enzyme activities, the activities of key enzymes showed good consistency with the methanogenesis pathway. Under 0.6 V perturbation, the activities of key enzymes increased, and the reactions in the catalyzed microorganisms accelerated, which had a high correlation with the changes in metabolic flux. The study also found that under voltage disturbance, the methane production fluxes of H2 reduction CO2 in the EAD system were 13.44% and 12.76% higher than those in the control group (5.97%), thereby improving the overall methane production capacity of the EAD system. -

-
表 1 EAD体系葡萄糖代谢过程包含的主要生化反应
Table 1. The main biochemical reactions included in the glucose metabolism process of the EAD system
NO. 反应式
EquationNO. 反应式
EquationR1 GLC+PEP ∀ G6P + Pyr R20 HPr + 2 H2O ∀ HAc + CO2 +3 H2 R2 NADH + Pyr ∀ HLa + NAD R21 HBu + 2 H2O ∀ 2 HAc + 2 H2 R3 Pyr + NADH ∀ NAD + HFo +AcCoA R22 HPr = HPr (ext) R4 2 Fd + Pyr + CoA ∀ CO2 + AcCoA + 2 FdH R23 EtOH+ HPr ∀ Hva (ext) + H2O R5 2 Fd + NADH = 2 FdH + NAD R24 HFo = HFo (ext) R6 NADPH + NAD ⇌ NADH + NADP R25 CO2 = CO2 (ext) R7 2 NADH = H2 + 2 NAD R26 CO2 + 4 H2 = CH4 + 2 H2O R8 2 FdH = H2 + 2 Fd R27 H2 = H2 (ext) R9 HLa = HLa (ext) R28 2 AcCoA + H2 ∀ PrOH(ext) +
2 CoA + CO2R10 HLa + NADH ∀ HPr + NAD R29 HAc ∀ CO2 + CH4 R11 HFo ∀ CO2 + H2 R30 HAc = HAc (ext) R12 AcCoA + ADP + iP ∀ATP + HAc + CoA R31 EtOH = EtOH (ext) R13 AcCoA + 2 NADH ∀ EtOH + CoA + 2 NAD R32 HBu + EtOH ∀ HCa (ext) + H2O R14 2 AcCoA + NADH ⇌ CoA + H2O + CroCoA + NAD R33 HBu = HBu (ext) R15 CroCoA + 2 Fd + NADH ∀ ButCoA + 2 FdH + NAD R34 HBu + 2 NADH ∀ BuOH(ext) + CoA + 2 NAD R16 CroCoA + NADH ∀ ButCoA + NAD R35 Pyr ⇌ Pyr (ext) R17 ButCoA + ADP + iP ∀ HBu + ATP + CoA R36 CH4 = CH4 (ext) R18 2 CO2 + 4 H2 ∀ HAc +2 H2O R37 2 H+ + 2 e- = H2 R19 EtOH + 2H2O ∀ 4 H+ + HAc 注:iP为磷酸根离子,CoA为辅酶A,H+为氢离子,e-为转移电子或者电解协助装置提供的电子. Note: iP is the phosphate ion, CoA is the coenzyme A, H+ is the hydrogen ion, and e- is the electron provided by the transfer electron or electrolysis assistance device. 表 2 EAD体系中关键代谢物的净生成速率①(mmol·L−1·h−1)
Table 2. Net formation rate of key metabolites in EAD system①(mmol·L−1·h−1)
代谢物
Metabolites对照组1.0 V
Control group 1.0 V扰动组0.6 V
Disturbance group 0.6 V扰动组1.4 V
Disturbance group 1.4 V葡萄糖② 1.1274 ± 0.0154 1.0958 ± 0.0055 1.1195 ± 0.0175 甲 酸 0.0032± 0.0000 0.0086 ± 0.0021 0.0093± 0.0026 乙 酸 0.7511 ± 0.1440 0.5229 ± 0.0770 0.7064 ± 0.1202 丙 酸 0.1061 ± 0.0096 0.0860 ± 0.0049 0.0902 ± 0.0023 丁 酸③ 0.4180± 0.0335 0.4218± 0.0391 0.4026 ± 0.0201 乳 酸 0.0107 ± 0.0046 0.0081 ± 0.0035 0.0128 ± 0.0056 丙酮酸 0.0156 ± 0.0037 0.0335± 0.0074 0.0261 ± 0.0171 戊 酸④ 0.0230 ± 0.0107 0.0107 ± 0.0046 0.0277 ± 0.0093 己 酸 0.0000 ± 0.0000 0.0000 ± 0.0000 0.0000 ± 0.0000 乙 醇 0.0173 ± 0.0014 0.0092 ± 0.0019 0.0160 ± 0.0067 丙 醇 0.0000 ± 0.0000 0.0000 ± 0.0000 0.0000 ± 0.0000 丁 醇 0.0000 ± 0.0000 0.0000 ± 0.0000 0.0000 ± 0.0000 CO2 0.2258 ± 0.0229 0.2255 ± 0.0376 0.2716 ± 0.0071 CH4 0.2947 ± 0.0127 0.4909 ± 0.0515 0.3924 ± 0.0293 H2 0.0281 ± 0.0231 0.0228 ± 0.0137 0.0039 ± 0.0024 注:表2中的实验数据①为电压扰动实验组的主要代谢物的净生成速率;葡萄糖②的测量值代表底物的净摄取速率;丁酸③为正丁酸和异丁酸的总和,戊酸④为正戊酸和异戊酸的总和. Note: The experimental data① in Table 2 are the net production rates of major metabolites in the voltage disturbance group; The measured value of glucose② represents the net uptake rate of substrate; Butyrate③ is the sum of n-butyrate and isobutyrate, valerate④ is the sum of n-valerate and isovalerate. 表 3 代谢途径中关键酶活性水平的变化
Table 3. Changes in activity levels of key enzymes in metabolic pathways
酶名称
Enzyme对照组1.0 V
Control group 1.0 V扰动组0.6 V
Disturbance group 0.6 V扰动组1.4 V
Disturbance group 1.4 V单位
UnitADP/ATP转运酶 239.28±23.54 337.04±7.88 344.23±7.54 IU·L−1 磷酸转乙酰酶(PTA) 99.78±6.39 115.69±11.32 97.69±7.50 U·L−1 乙酸激酶(AK) 106.26±24.23 151.38±28.81 120.25±28.93 U·L−1 磷酸转丁酰酶(PTB) 310.19±1.29 352.19±12.66 338.04±22.08 U·L−1 丁酸激酶(BK) 52.59±6.63 50.92±0.90 36.08±4.89 U·L−1 辅酶M(CoM) 173.84±17.97 191.01±23.53 176.85±12.82 U·L−1 甲基丙二酰CoA变位酶(MCM) 58.17±4.06 51.71±2.71 54.20±3.09 U·L−1 辅酶F420(CoF420) 40.38±2.22 43.72±4.37 36.86±6.48 IU·L−1 辅酶F430(CoF430) 16.56±2.05 19.29±1.55 22.71±2.14 IU·L−1 -
[1] ZHANG Z Y, SONG Y, ZHENG S J, et al. Electro-conversion of carbon dioxide (CO2) to low-carbon methane by bioelectromethanogenesis process in microbial electrolysis cells: The current status and future perspective [J]. Bioresource Technology, 2019, 279: 339-349. doi: 10.1016/j.biortech.2019.01.145 [2] LIN C B, WU P, LIU Y D, et al. Enhanced biogas production and biodegradation of phenanthrene in wastewater sludge treated anaerobic digestion reactors fitted with a bioelectrode system [J]. Chemical Engineering Journal, 2019, 365: 1-9. doi: 10.1016/j.cej.2019.02.027 [3] PARK J, LEE B, TIAN D, et al. Bioelectrochemical enhancement of methane production from highly concentrated food waste in a combined anaerobic digester and microbial electrolysis cell [J]. Bioresource Technology, 2018, 247: 226-233. doi: 10.1016/j.biortech.2017.09.021 [4] TIAN T, QIAO S, YU C, et al. Bio-electrochemically assisting low-temperature anaerobic digestion of low-organic strength wastewater [J]. Chemical Engineering Journal, 2018, 335: 657-664. doi: 10.1016/j.cej.2017.11.016 [5] XIAO B Y, CHEN X, HAN Y P, et al. Bioelectrochemical enhancement of the anaerobic digestion of thermal-alkaline pretreated sludge in microbial electrolysis cells [J]. Renewable Energy, 2018, 115: 1177-1183. doi: 10.1016/j.renene.2017.06.043 [6] XU S Y, ZHANG Y C, LUO L W, et al. Startup performance of microbial electrolysis cell assisted anaerobic digester (MEC-AD) with pre-acclimated activated carbon [J]. Bioresource Technology Reports, 2019, 5: 91-98. doi: 10.1016/j.biteb.2018.12.007 [7] HO D, JENSEN P, BATSTONE D. Effects of temperature and hydraulic retention time on acetotrophic pathways and performance in high-rate sludge digestion [J]. Environmental Science & Technology, 2014, 48(11): 6468-6476. [8] CHEN M, LIU S J, YUAN X F, et al. Methane production and characteristics of the microbial community in the co-digestion of potato pulp waste and dairy manure amended with biochar [J]. Renewable Energy, 2021, 163: 357-367. doi: 10.1016/j.renene.2020.09.006 [9] RAFIEENIA R, PIVATO A, SCHIEVANO A, et al. Dark fermentation metabolic models to study strategies for hydrogen consumers inhibition [J]. Bioresource Technology, 2018, 267: 445-457. doi: 10.1016/j.biortech.2018.07.054 [10] GONZÁLEZ-CABALEIRO R, LEMA J M, RODRÍGUEZ J. Metabolic energy-based modelling explains product yielding in anaerobic mixed culture fermentations [J]. PLoS One, 2015, 10(5): e0126739. doi: 10.1371/journal.pone.0126739 [11] VALLINO J J, STEPHANOPOULOS G. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction [J]. Biotechnology and Bioengineering, 1993, 41(6): 633-646. doi: 10.1002/bit.260410606 [12] GONZALEZ-GARCIA R A, AISPURO-CASTRO R, SALGADO-MANJARREZ E, et al. Metabolic pathway and flux analysis of H2 production by an anaerobic mixed culture [J]. International Journal of Hydrogen Energy, 2017, 42(7): 4069-4082. doi: 10.1016/j.ijhydene.2017.01.043 [13] GAO H J, DU G C, CHEN J. Analysis of metabolic fluxes for hyaluronic acid (HA) production by Streptococcus zooepidemicus [J]. World Journal of Microbiology and Biotechnology, 2006, 22(4): 399-408. doi: 10.1007/s11274-005-9047-7 [14] KÖPKE M, HELD C, HUJER S, et al. Clostridium ljungdahlii represents a microbial production platform based on syngas [J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(29): 13087-13092. doi: 10.1073/pnas.1004716107 [15] LESKOVAC V, TRIVIĆ S, PERIČIN D. The three zinc-containing alcohol dehydrogenases from baker's yeast, Saccharomyces cerevisiae [J]. FEMS Yeast Research, 2002, 2(4): 481-494. [16] KUNJI E R S, ALEKSANDROVA A, KING M S, et al. The transport mechanism of the mitochondrial ADP/ATP carrier [J]. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 2016, 1863(10): 2379-2393. doi: 10.1016/j.bbamcr.2016.03.015 [17] LIU H Z, ZHANG Y R, YANG S X, et al. Introducing electrolysis to enhance anaerobic digestion resistance to acidification [J]. Bioprocess and Biosystems Engineering, 2022, 45(3): 515-525. doi: 10.1007/s00449-021-02675-8 [18] MOJUMDAR A, UPADHYAY A K, RAINA V, et al. A simple and rapid colorimetric method for the estimation of chitosan produced by microbial degradation of chitin waste [J]. Journal of Microbiological Methods, 2019, 158: 66-70. doi: 10.1016/j.mimet.2019.02.001 [19] SHENG W, YANG L, WANG J P, et al. Development of an enzyme-linked immunosorbent assay for the detection of gentamycin residues in animal-derived foods [J]. LWT - Food Science and Technology, 2013, 50(1): 204-209. doi: 10.1016/j.lwt.2012.05.028 [20] CHOI J H, LIM Y T, OH B K. Development of colorimetric enzyme-ball for signal amplification of enzyme-linked immunosorbent assay [J]. Science of Advanced Materials, 2014, 6(11): 2572-2576. doi: 10.1166/sam.2014.2225 [21] VARMA A, PALSSON B O. Metabolic flux balancing: Basic concepts, scientific and practical use [J]. Bio/Technology, 1994, 12(10): 994-998. doi: 10.1038/nbt1094-994 [22] KAUFFMAN K J, PRAKASH P, EDWARDS J S. Advances in flux balance analysis [J]. Current Opinion in Biotechnology, 2003, 14(5): 491-496. doi: 10.1016/j.copbio.2003.08.001 [23] DYKSTRA C M, PAVLOSTATHIS S G. Zero-valent iron enhances biocathodic carbon dioxide reduction to methane [J]. Environmental Science & Technology, 2017, 51(21): 12956-12964. [24] WANG Y Y, ZHANG Y L, WANG J B, et al. Effects of volatile fatty acid concentrations on methane yield and methanogenic bacteria [J]. Biomass and Bioenergy, 2009, 33(5): 848-853. doi: 10.1016/j.biombioe.2009.01.007 [25] FACHET M, WITTE C, FLASSIG R J, et al. Reconstruction and analysis of a carbon-core metabolic network for Dunaliella salina [J]. BMC Bioinformatics, 2020, 21(1): 1. doi: 10.1186/s12859-019-3325-0 [26] LEE H S, KRAJMALINIK-BROWN R, ZHANG H S, et al. An electron-flow model can predict complex redox reactions in mixed-culture fermentative BioH2: Microbial ecology evidence [J]. Biotechnology and Bioengineering, 2009, 104(4): 687-697. [27] CHAGANTI S R, KIM D H, LALMAN J A. Flux balance analysis of mixed anaerobic microbial communities: Effects of linoleic acid (LA) and pH on biohydrogen production [J]. International Journal of Hydrogen Energy, 2011, 36(21): 14141-14152. doi: 10.1016/j.ijhydene.2011.04.161 [28] SI B C, YANG H, HUANG S J, et al. An innovative multistage anaerobic hythane reactor (MAHR): Metabolic flux, thermodynamics and microbial functions [J]. Water Research, 2020, 169: 115216. doi: 10.1016/j.watres.2019.115216 [29] WANG Y L, WANG D B, CHEN F, et al. Effect of triclocarban on hydrogen production from dark fermentation of waste activated sludge [J]. Bioresource Technology, 2019, 279: 307-316. doi: 10.1016/j.biortech.2019.02.016 [30] CHEN Y G, LIU H, ZHENG X, et al. New method for enhancement of bioenergy production from municipal organic wastes via regulation of anaerobic fermentation process [J]. Applied Energy, 2017, 196: 190-198. doi: 10.1016/j.apenergy.2017.01.100 [31] WANG C, WANG C Q, LIU J Y, et al. Role of magnetite in methanogenic degradation of different substances [J]. Bioresource Technology, 2020, 314: 123720. doi: 10.1016/j.biortech.2020.123720 [32] ZHOU Q, LIU Y, YUAN W Q. Kinetic modeling of butyric acid effects on butanol fermentation by Clostridium saccharoperbutylacetonicum [J]. New Biotechnology, 2020, 55: 118-126. doi: 10.1016/j.nbt.2019.10.004 [33] YU Y, SHAO M Y, LI D, et al. Construction of a carbon-conserving pathway for glycolate production by synergetic utilization of acetate and glucose in Escherichia coli [J]. Metabolic Engineering, 2020, 61: 152-159. doi: 10.1016/j.ymben.2020.06.001 [34] SIKORA A, DETMAN A, MIELECKI D, et al. Searching for metabolic pathways of anaerobic digestion: A useful list of the key enzymes[M]. 2018. [35] ZHU Y, YANG S T. Effect of pH on metabolic pathway shift in fermentation of xylose by Clostridium tyrobutyricum [J]. Journal of Biotechnology, 2004, 110(2): 143-157. doi: 10.1016/j.jbiotec.2004.02.006 [36] JO J H, KIM W. Carbon material distribution and flux analysis under varying glucose concentrations in hydrogen-producing Clostridium tyrobutyricum JM1 [J]. Journal of Biotechnology, 2016, 228: 103-111. doi: 10.1016/j.jbiotec.2016.04.051 -




 下载:
下载: