-
汞(Hg)是一种能随大气进行长距离传输的全球性环境污染物[1 − 2]. 自然环境中的汞可分为无机汞(汞单质、一价汞和二价汞等)和有机汞(甲基汞、乙基汞和苯基汞等). 其中,甲基汞(MeHg)由于强的神经毒性和生物富集性而引起人们的广泛关注[3]. 无机汞的甲基化是汞生物地球化学循环中的重要环节,同时威胁着人类的健康.
一般认为,厌氧微生物作用是驱动无机汞转化为甲基汞最主要的方式[4 − 7]. HgcAB基因调控微生物的汞甲基化过程,是汞甲基化微生物的标志性基因[8]. 根据hgcAB基因在微生物中的分布,将汞甲基化微生物分为5个进化枝,包括δ-变形菌门的3类(硫酸盐还原菌、铁还原菌、互营杆菌)、厚壁菌门和部分古菌(产甲烷菌)[2,9]. 过去很长一段时间,关于微生物汞甲基化的研究大多以硫酸盐还原菌和铁还原菌展开. 近年来的研究表明,产甲烷菌在汞甲基化过程中具有重要地位:(1)在沉积物、水体和土壤等环境中,产甲烷菌与汞甲基化过程密切相关[10 − 12]. 部分区域中,汞甲基化关键基因(hgcAB基因)在产甲烷菌中的基因丰度显著高于其他汞甲基化微生物[13 − 15];(2)在实验室纯培养体系中,多株产甲烷菌具有与硫酸盐还原菌、铁还原菌相当甚至更高的汞甲基化效率[9, 16 − 17];(3)进化分析显示,hgcAB基因和汞甲基化途径的钴铁硫蛋白起源于产甲烷菌,产甲烷菌可能是最早进行汞甲基化的微生物[18].
目前,尽管部分关于微生物汞甲基化的综述中对产甲烷菌汞甲基化的研究进展进行简单归纳[6 − 7, 19 − 20],但缺乏对产甲烷菌汞甲基化作用的系统总结. 同时,关于微生物汞甲基化途径的研究主要集中在硫酸盐还原菌和铁还原菌,缺乏对产甲烷菌汞甲基化途径的探讨. 本文在前人研究的基础上,系统总结了产甲烷菌在原位环境、纯培养和共培养体系中对汞甲基化的作用. 从生物代谢的角度,对产甲烷菌汞甲基化的途径进行探讨,提出产甲烷途径是产甲烷菌汞甲基化可能的途径之一,以期为微生物汞甲基化的研究提供新的方向和思路.
-
近年来,产甲烷菌对汞甲基化的显著贡献引起了人们的广泛关注. 随着hgcAB基因的发现和基因组学、蛋白质组学等方法的发展[20 − 21],产甲烷菌的汞甲基化作用在沉积物、水体和土壤等多种环境中被发现(表1),在部分地区的河口沉积物和湖泊周丛生物中,产甲烷菌主导甲基汞的生成.
厌氧沉积物普遍被认为是硫酸盐还原菌和铁还原菌汞甲基化的主要场所[36 − 38],而最近研究表明,产甲烷菌在多个河流、湖泊和海洋沉积物的汞甲基化过程中扮演重要角色[10, 13, 22]. 由黏土、金属碳酸盐等矿物组成的厌氧沉积物为细菌、古菌和藻类等提供了良好的微环境和栖息地,有利于甲基汞的生成. 通过16S rRNA基因测序和实时荧光定量PCR等方法,检测到厌氧沉积物中的产甲烷菌主要属于甲烷微菌纲(Methanomicrobia). 在高硫酸盐含量的圣哈辛托河(San Jacinto River)的河口沉积物中,产甲烷菌抑制剂2-溴乙烷磺酸钠(BES)几乎完全抑制了甲基汞的生成,相较于不加BES,甲基汞浓度减少了12倍[10]. 同时检测到产甲烷菌甲烷生成和汞甲基化的关键基因——mcrA和hgcA,其中的产甲烷菌主要属于甲烷八叠球菌科(Methanosarcinaceae)[10]. 在对橡树岭东部河流沉积物的分析中,产甲烷菌的丰度是其他汞甲基化微生物的2—5倍,甲烷鬃菌科(Methanosaetaceae)、甲烷八叠球菌科(Methanosarcinaceae)和甲烷球菌科(Methanococcaceae)是该河流沉积物中主要的产甲烷菌[13]. 在圣莫里斯河(Saint Maurice River)和瑞典北部的湖泊沉积物中,产甲烷菌中的甲烷绳菌属(Methanolinea)、甲烷螺菌属(Methanospirillum)、马赛球菌属(Methanomassiliicoccus)和Methanoregula对汞甲基化具有一定的贡献[23 − 24]. 另外,在多个海洋沉积物中检测到马赛球菌属(Methanomassiliicoccus)、甲烷螺菌属(Methanospirillum)、甲烷叶菌属(Methanolobus)和甲烷泡菌属(Methanofollis)等的hgcA基因[18].
相较于沉积物,河流、湖泊和海洋等水体中的甲基汞更容易被初级消费者吸收利用,并通过食物链不断富集、放大. 研究表明,产甲烷菌在水生周丛植物生物膜中主导甲基汞的生成[11],同时参与富营养水体[14]和海水[25 − 26]中的汞甲基化过程. 在圣劳伦斯河(Saint. Lawrence River)的水生周丛植物生物膜中,通过同位素标记、生物抑制剂和16S rRNA基因测序相结合的方法,发现产甲烷菌是汞甲基化过程中的主要贡献者[11]. 在添加产甲烷菌抑制剂BES的情况下,甲基汞的生成完全被抑制;而在添加硫酸盐还原抑制剂(钼酸盐)时,汞甲基化效率不仅没有降低,反而提升了45倍,产甲烷菌在整个微生物群落的占比从21.2%增加到33.3%. 其中的产甲烷菌包括甲烷杆菌目(Methanobacteriales)、甲烷球菌目(Methanococcales)和甲烷八叠球菌目(Methanosarcinales)[11]. 在对富营养化水体的研究中,产甲烷菌的hgcA基因丰度高于硫酸盐还原菌,藻类有机质的输入显著促进产甲烷菌的活性和hgcA基因丰度,产甲烷菌可能是导致富营养水体甲基汞增加的主要原因[14]. 在太平洋和南极地区的海水中,马赛球菌属(Methanomassiliicoccus)和甲烷微菌纲(Methanomicrobia)的产甲烷菌分别参与到微生物汞甲基化过程中[25 − 26].
土壤是大气汞沉降和微生物活动的主要场所. 在稻田土壤[12, 15, 27 − 28, 39]、泥炭地[29]、湿地土壤[30, 31]和永久冻土[18, 32 − 35]等环境中,产甲烷菌对甲基汞的生成有一定的贡献,部分区域中,产甲烷菌的hgcAB基因丰度显著高于其他汞甲基化微生物. 稻田土壤作为典型的季节性水驱湿地生态系统,在淹水状态下,好氧微生物消耗土壤中的氧气,为无机汞甲基化提供了良好的厌氧条件[40 − 41]. 金属矿区附近的稻田土壤作为受汞污染的典型土壤被广泛研究. 在我国万山矿区和凤凰矿区附近的稻田土壤中,产甲烷菌的hgcAB基因丰度(57%)显著高于硫酸盐还原菌(<3%)和铁还原菌(34%)[15]. 在铜仁矿区附近的稻田土壤中,产甲烷菌的丰度与总汞、甲基汞的浓度呈正相关[12]. 稻田土壤中的产甲烷菌主要属于甲烷微菌纲(Methanomicrobia)、甲烷球菌纲(Methanococci)、甲烷杆菌纲(Methanobacteria)和热源体纲(Thermoplasmata)[12, 15, 27]. 湿地作为土壤和地表水的混合区域,为汞甲基化微生物提供了厌氧的环境大量可分解的有机质. 在对瑞典北部典型泥炭地和湿地的研究中,产甲烷菌的mcrA和hgcA基因与甲基汞的生成量成显著正相关,其中的产甲烷菌包括甲烷八叠球菌科(Methanosarcinales)、甲烷杆菌科(Methanobacteriaceae)、马赛球菌科(Methanomassiliicoccaceae)和Methanoregulaceae[29, 31]. 另外,在青藏高原、博南扎河和阿拉斯加州等地区的永久冻土中,甲基汞的生成和产甲烷菌的hgcAB基因丰度、甲烷的生成有关[18, 32 − 35].
-
产甲烷菌属于严格厌氧菌,生长缓慢,对氧极其敏感,通常认为产甲烷菌在氧化还原电位低于-350 mV时才能正常生长[42 − 43]. 同时,产甲烷菌只能利用甲酸钠、乙酸钠、H2、CO2、甲醇、甲胺和甲硫醇等简单的物质进行能量代谢. 这些条件极大的限制了产甲烷菌纯培养条件下的汞甲基化研究.
最早关于产甲烷菌纯培养体系下的汞甲基化研究可以追溯到20世纪60年代,产甲烷菌Methanobacterium bryantii在H2和CO2的混合气中培养,其细胞提取物能够快速地将Hg2+转化为甲基汞[44 − 45]. 有趣的是,后来的研究并没有在M. bryantii中检测到微生物汞甲基化的关键基因—hgcAB基因. 目前,通过基因测序和序列对比等方法,共在18株产甲烷菌中发现hgcAB基因的同源序列,其中8株菌被证明能将无机汞转化为甲基汞[7, 16, 18],它们的分类、培养条件和甲基化效率如表2所示. 除Methanomassiliicoccus luminyensis B10属于热源体纲(Thermoplasmata)的马赛球菌目(Methanomassiliicoccales)外,其余7株汞甲基化产甲烷菌均属于甲烷微菌纲(Methanomicrobia),它们分布在甲烷微菌目(4株)、甲烷八叠球菌目(2株)和甲烷胞菌目(1株). 马赛球菌目的Methanomassiliicoccus luminyensis B10和甲烷八叠球菌目的Methanolobus tindarius、Methanomethylovorans hollandica为甲基营养型产甲烷菌[46 − 48],利用甲醇、甲胺、甲硫醇和甲硫醚等物质进行代谢. 其余属于甲烷微菌目和甲烷胞菌目的5株菌为氢营养型产甲烷菌,以CO2、H2或甲酸进行能量代谢[49]. 8株被证实的汞甲基化产甲烷菌中,有4株菌的汞甲基化效率大于10%[9, 16]. 甲烷微菌目的Methanospirillum hungatei JF-1是第一个在纯培养条件下被证实的汞甲基化产甲烷菌[17],同时也是甲烷螺菌科(Methanospirillaceae)中最早分离和测定出完整基因组的产甲烷菌[50],最高能将64.2%的Hg2+转化为甲基汞[16].
产甲烷菌对氧化还原电位要求极高,还原剂是降低培养体系氧化还原电位的方法之一. 硫化物、次氮基三乙酸钛(Ti-NTA)和半胱氨酸作为汞甲基化微生物培养过程中3种常见的还原剂,对汞甲基化过程有重要影响. 硫化物对汞甲基化的影响呈现低浓度促进,高浓度抑制的效果. 低浓度的硫化物有利于中性硫化汞配合物(HgS0(aq))的形成,HgS0(aq)在被动运输的跨膜传递过程中更容易被吸收,一定程度上促进了微生物汞甲基化[51]. 而在高浓度的硫化物中,S2-与Hg2+更容易生成不溶性的HgS沉淀(HgS(s)),降低了Hg2+的生物可利用性,从而抑制了微生物汞甲基化过程[52 − 54]. 相比于Na2S,次氮基三乙酸钛(Ti-NTA)更适合产甲烷菌的培养,Methanospirillum hungatei JF-1在Ti-NTA中的甲基化效率是Na2S的15倍[17]. 半胱氨酸不仅为产甲烷菌的生长提供还原性的硫源,促进微生物的生长,而且能与Hg2+形成配合物,提高微生物汞甲基化效率[16].
-
在厌氧环境中,电子受体更容易耗尽. 普遍认为,厌氧环境中的汞甲基化过程由多种微生物相互作用、共同驱动[29]. 相较于单一纯菌的培养,多菌的共培养体系能更好地模拟实际环境中的汞甲基化过程,进一步探究微生物相互作用对汞甲基化的影响.
在硫酸盐和铁离子含量较低的厌氧环境中,产甲烷菌与其他微生物之间的相互作用可能是甲基汞的主要来源. 研究表明,在没有硫酸盐存在的共培养体系中,汞甲基化产甲烷菌Methanospirillum hungatei JF-1能够显著促进硫酸盐还原菌Desulfovibrio africanus和Desulfovibrio desulfuricans ND132的汞甲基化,相较于单一纯菌培养体系,汞甲基化效率提高了2—9倍[59]. Methanospirillum hungatei JF-1在与互营菌Syntrophobacter wolinii共培养的过程中,汞甲基化效率提升了2倍[59]. 另外,非汞甲基化产甲烷菌Methanococcus maripaludis同样能够促进甲基汞的生成[18]. 在乳酸-碳酸盐培养体系中,单独培养M. maripaludis和D. desulfuricans LS均未观察到明显的汞甲基化现象. 而在共同培养体系中,2.6%的无机汞转化为甲基汞[60]. 这也表明非汞甲基化微生物同样在共生环境中发挥作用.
共培养体系中,产甲烷菌与其他微生物之间广泛的物质交换和能量传递可能是汞甲基化效率显著增强的原因[60]. 其中,硫酸盐还原菌、互营菌等δ变形菌能将丙酸、丁酸等中间体转化为乙酸、甲酸和H2. 而产甲烷菌能够利用乙酸、甲酸和H2进行代谢,同时伴随着能量的产生,该过程促进了微生物的生长和代谢[60 − 63].
-
产甲烷菌在汞甲基化过程中发挥重要作用,目前关于产甲烷菌汞甲基化作用途径和机制的研究还很少. 系统发育表明产甲烷菌可能是最古老的汞甲基化微生物,产甲烷菌汞甲基化途径的研究对于理解微生物汞甲基化机制和减少环境中甲基汞的生成具有重要意义. 早期研究发现,产甲烷菌提取物中的甲基钴胺素能为汞甲基化提供甲基[45],甲基钴胺素也成为微生物汞甲基化机制研究中的关键物质. 基于以往的研究和产甲烷菌的代谢特点,本文对产甲烷菌中可能的汞甲基化途径进行讨论.
-
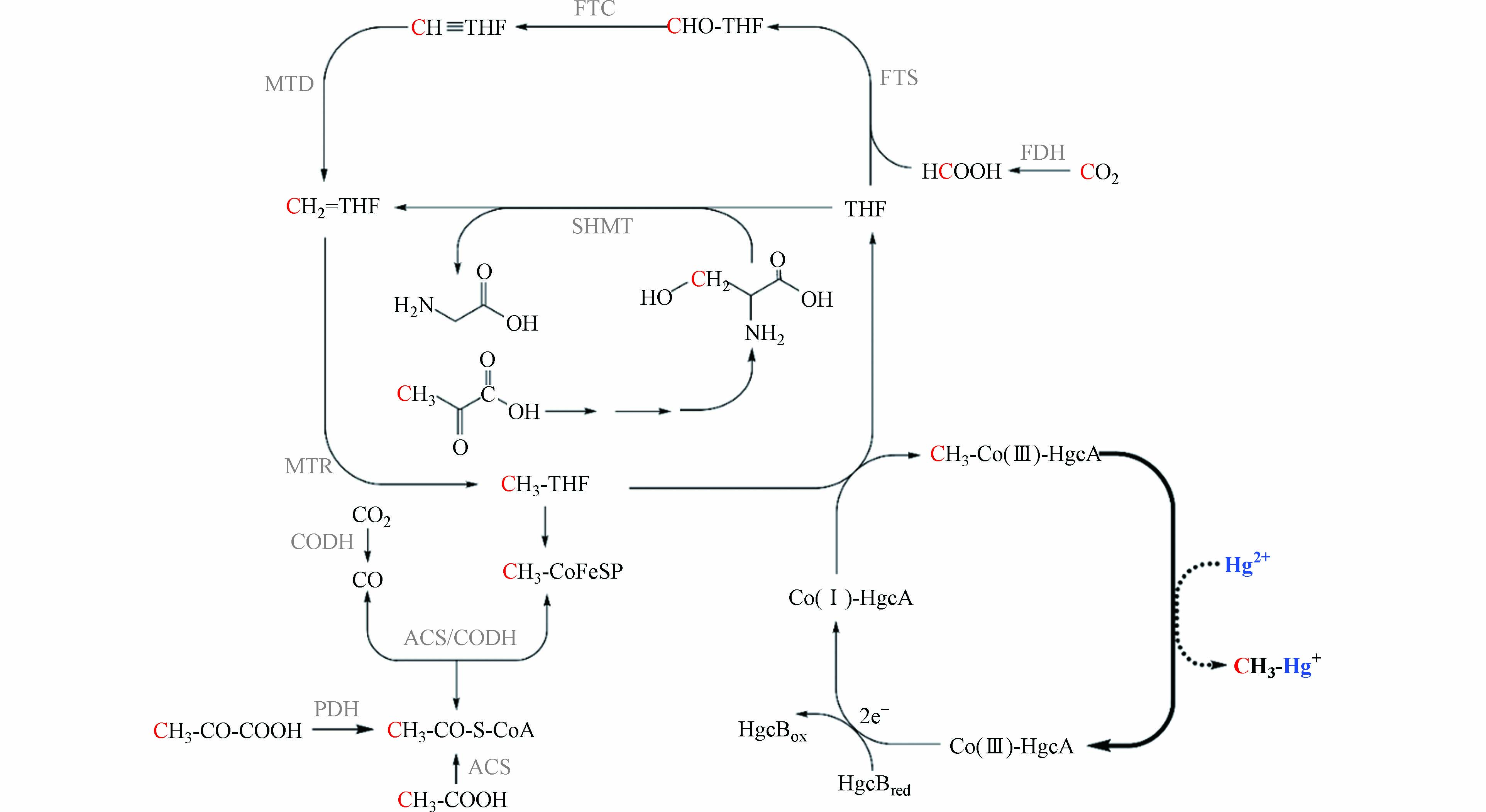
乙酰辅酶A途径是广泛存在于古菌和厌氧细菌中的古老代谢途径[64 − 65],也是产甲烷菌一碳代谢的方式之一[66]. 产甲烷菌等微生物通过乙酰辅酶A途径将CO2转化为乙酸等较为复杂的有机化合物,该过程被认为是生命出现早期微生物合成所需物质的主要途径[67 − 69]. 如图1所示,CO2或甲酸与四氢叶酸(THF)结合,在一系列还原酶的催化作用下转化为甲基钴胺素,甲基钴胺素与CO在乙酰辅酶A合成酶(ACS)的作用下合成乙酰辅酶A[64].
乙酰辅酶A途径是研究较为广泛的微生物汞甲基化途径. 系统发育分析表明,产甲烷菌是最早出现乙酰辅酶A途径中钴铁硫蛋白的汞甲基化微生物,硫酸盐还原菌和铁还原菌中的钴铁硫蛋白可能来源于产甲烷菌[18]. 氢营养型产甲烷菌可能是最先利用乙酰辅酶A途径进行代谢的汞甲基化微生物[18, 68]. 但目前并没有直接验证产甲烷菌通过乙酰辅酶A途径进行汞甲基化的报道. 研究者通过14C同位素标记的方法,发现硫酸盐还原菌Desulfovibrio desulfuricans LS通过乙酰辅酶A途径进行汞甲基化[70]. 2013年,汞甲基化关键基因——hgcAB基因被发现,同时认为hgcA基因编码的类钴啉蛋白将甲基四氢叶酸上的甲基转移给汞[8]. 基于产甲烷菌提取物中甲基钴胺素在汞甲基化中的作用[45]以及hgcAB基因在多个产甲烷菌中的分布[18],产甲烷菌很可能同样通过乙酰辅酶A途径进行汞甲基化. 乙酰辅酶A途径的甲酸、丝氨酸和丙酮酸是甲基汞生成的潜在甲基供体[70],如图1所示,它们通过不同的途径将甲基转移给甲基四氢叶酸,并在hgcAB基因的参与下完成汞甲基化过程. 其中,甲酸通过叶酸分支转化为甲基四氢叶酸;丝氨酸的C-3在丝氨酸羟甲基转移酶(SHMT)的催化下转移给四氢叶酸,并在叶酸循环中转化为甲基四氢叶酸;丙酮酸既可以通过丝氨酸将甲基传递[70 − 71],又可以在丙酮酸脱氢酶(PDH)的作用下将甲基转移给乙酰辅酶A,裂解生成甲基四氢叶酸. 未来,关于乙酰辅酶A途径在产甲烷菌汞甲基化中的作用和甲基转移过程还需要进一步的确认.
-
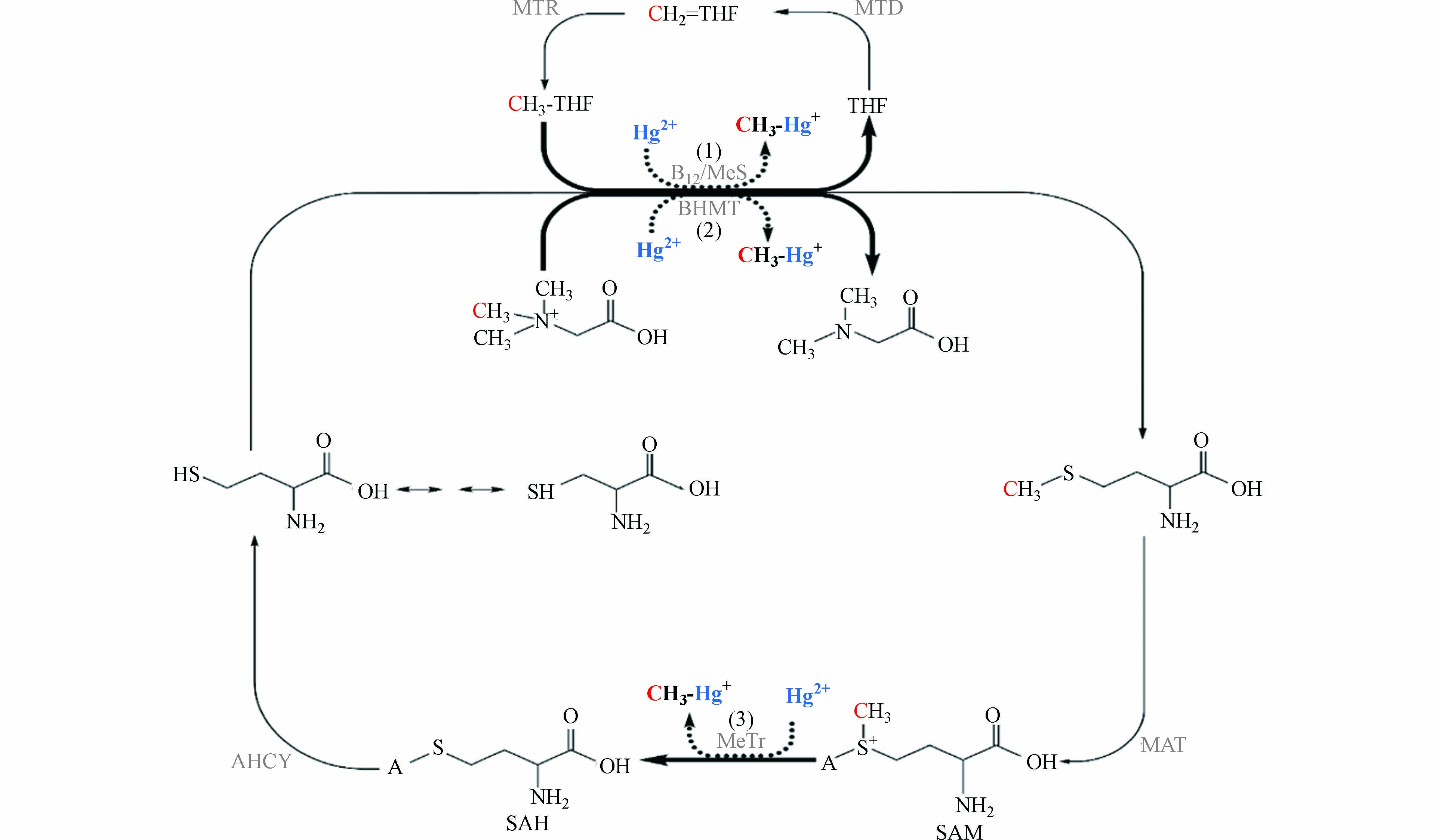
甲硫氨酸循环与叶酸循环密切相关[72 − 74],同样属于微生物的一碳代谢过程. 如图2所示,甲基四氢叶酸或甜菜碱为同型半胱氨酸提供甲基,生成甲硫氨酸[75 − 76]. 之后,甲硫氨酸在甲硫氨酸腺苷甲基转移酶(MAT)的催化下,生成S-腺苷甲硫氨酸(SAM). SAM作为重要的甲基供体参与多种物质的甲基化过程[77 − 78],脱去甲基的SAM转化为S-腺苷同型甲硫氨酸(SAH). 最后,SAH在腺苷同型半胱氨酸酶(AHCY)的作用下生成同型半胱氨酸,完成甲硫氨酸循环过程.
早在1971年,甲硫氨酸循环就被提出是可能的汞甲基化途径. 研究发现,在真菌Neurospora crassa中,汞抗性与甲硫氨酸合成相关,并且同型半胱氨酸和甲硫氨酸的加入能够促进汞甲基化[79]. 一些缺乏乙酰辅酶A活性的非完全氧化硫酸盐还原菌同样能进行汞甲基化,其中,甲硫氨酸循环可能参与了汞甲基化过程[80 − 81]. 目前,还没有关于甲硫氨酸循环途径与产甲烷菌汞甲基化关系的研究. 基于前人的研究和甲硫氨酸代谢的特点,推测在一些没有完整乙酰辅酶A途径的产甲烷菌中,甲硫氨酸途径是可能的汞甲基化途径. 甲硫氨酸途径中存在3处可能的汞甲基化位点:(1)甲基四氢叶酸上的甲基在钴胺素的参与下转移给汞;(2)甜菜碱上的甲基在甲基转移酶的作用下转移给汞;(3)SAM上的甲基在钴胺素或其他亲核受体的参与下转移给汞[82]. 其中,位点(1)与乙酰辅酶A途径存在相似之处,均为甲基四氢叶酸上的甲基转移给Cob(Ⅰ)alamin,生成CH3-Cob(Ⅲ)alamin. 值得注意的是,Cob(Ⅰ)alamin会偶然生成Cob(Ⅱ)alamin,SAM能为Cob(Ⅱ)alamin提供甲基,在甲硫氨酸合成还原酶的作用下生成CH3-Cob(Ⅲ)alamin[83 − 84]. 位点(2)(3)中的甜菜碱和SAM提供带正电的甲基,从化学反应的角度来看,汞甲基化过程需要带负电的甲基[85 − 86],因此,甜菜碱和SAM上的甲基可能需要亲核受体作为中间体将甲基进行转移[87].
-
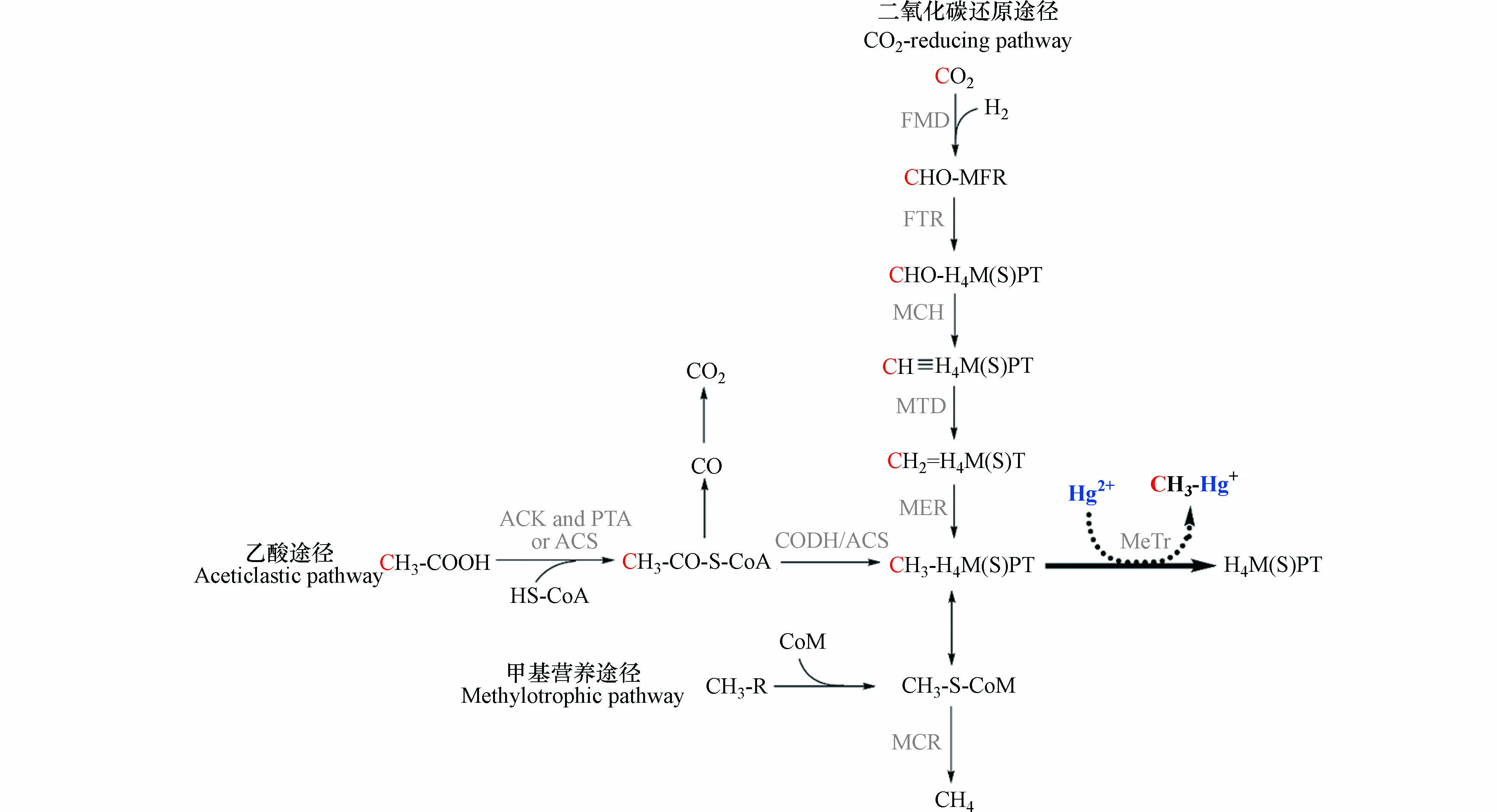
产甲烷途径是产甲烷菌特有的能量来源和一碳代谢途径[49]. 根据利用底物的不同,可将产甲烷途径分为3类(图3):还原CO2途径、乙酸途径和甲基营养途径. 还原CO2途径是大多数产甲烷菌的代谢方式,该过程以H2或甲酸作为电子供体,CO2作为电子受体,CO2在甲烷呋喃、四氢甲烷蝶呤和辅酶M等一碳载体的传递下,逐步还原生成甲烷. 甲基营养途径以甲醇、甲胺(一甲胺、二甲胺、三甲胺、四甲胺)和甲基硫化物(甲硫醇、甲硫醚)等为底物,在不同的底物特异性甲基转移酶作用下,将甲基转移到辅酶M上,之后还原生成甲烷. 乙酸途径首先将乙酸转化为乙酰辅酶A,并在一氧化碳脱氢酶/乙酰辅酶A合成酶复合体(CODH/ACS)的催化作用下生成甲基四氢八叠蝶呤和CO2,之后将甲基转移给辅酶M并生成甲烷[49, 88].
研究发现,汞甲基化产甲烷菌Methanospirillum hungatei缺乏催化CO羰基化的酶[89],Methanofollis liminatans、Methanocorpusculum bavaricum和Methanocella paludicola没有乙酰辅酶A途径的钴铁硫蛋白[18],而它们在实验室纯培养中,均能将无机汞转化为甲基汞. 因此,乙酰辅酶A途径并非产甲烷菌汞甲基化的唯一途径. 产甲烷途径作为古老的一碳代谢途径,与乙酰辅酶A途径存在物质组成和进化上的相似性,是可能的汞甲基化途径. 首先,还原CO2途径与乙酰辅酶A途径的叶酸分支类似,乙酸途径中包含了乙酰辅酶A的分解过程. 其次,甲基四氢甲烷蝶呤和甲基四氢八叠蝶呤是甲基四氢叶酸的类似物[90],三者与甲基结合的蝶呤环高度相似. 在进化上,产甲烷途径可能是从乙酰辅酶A途径进化而来[66]. 因此,甲基四氢甲烷蝶呤和甲基四氢八叠蝶呤可能与甲基四氢叶酸的作用相同,作为直接甲基供体参与汞甲基化过程.
-
综上所述,产甲烷菌作为古老的汞甲基化微生物,不仅在原位环境中对甲基汞的生成至关重要,而且在实验室纯培养和共培养过程中具有较高的汞甲基化效率. 因此,产甲烷菌对于汞的生物地球化学循环和微生物汞甲基化机制的研究具有重要意义. 乙酰辅酶A途径、甲硫氨酸途径和产甲烷途径是3种可能的产甲烷菌汞甲基化途径,这3种途径均属于微生物的一碳代谢过程,表明微生物汞甲基化过程与微生物的一碳代谢极大可能存在功能上的关联性. 因此,甲基汞的生物合成可能更容易发生在一碳底物浓度较高和一碳代谢较旺盛的环境,这类环境应作为甲基汞污染防治重点关注的区域.
目前关于产甲烷菌汞甲基化的过程和机理认识还存在较大不足,仍有许多亟待解决的问题. (1)不同环境中产甲烷菌对甲基汞生成的贡献. 产甲烷菌参与多种环境的汞甲基化过程,但在每种环境中产甲烷菌对汞甲基化的具体贡献尚不清楚. (2)规范产甲烷菌汞甲基化研究的培养条件,建立产甲烷菌汞甲基化研究的模式菌. 建立稳定的产甲烷菌培养体系和产甲烷菌汞甲基化研究的模式菌对于未来的研究十分有必要. (3)探究产甲烷菌汞甲基化的分子机制. 一方面,在实验室纯培养和共培养体系中,利用同位素标记、代谢组学分析和特定代谢通路阻断等方法,研究产甲烷菌汞甲基化的生化分子代谢路径;另一方面,通过对实际环境中汞甲基化过程的研究,进一步阐明微生物汞甲基化的机制.
产甲烷菌中汞甲基化研究进展与展望
Research progress and prospect of mercury methylation in methanogens
-
摘要: 微生物作用是无机汞转化为甲基汞最主要的方式. 其中,硫酸盐还原菌和铁还原菌被认为是主要的汞甲基化微生物,近年来的研究表明,产甲烷菌在汞甲基化过程中同样发挥重要作用. 产甲烷菌参与沉积物、水体和土壤等多种环境中的汞甲基化过程,并在部分环境中主导甲基汞的生成;在实验室纯培养体系中,多个产甲烷菌能将10%以上的无机汞转化为甲基汞. 产甲烷菌对于甲基汞的生成至关重要,然而目前缺乏对产甲烷菌汞甲基化作用的系统总结. 本文分别从原位环境、纯培养和共培养的角度,详细阐述了产甲烷菌在汞甲基化中的作用. 在此基础上,对产甲烷菌可能的汞甲基化途径进行探讨,提出汞甲基化过程与一碳代谢极大可能存在功能上的关联性,并展望了后续研究的方向和重点.Abstract: Microbial transformation is the most important way to convert inorganic mercury to methylmercury. Among them, sulfate-reducing bacteria and iron-reducing bacteria are considered to be the primary mercury methylation microorganisms. In recent years, several studies have shown that methanogens also play an important role in mercury methylation. Methanogens are involved in the process of mercury methylation in a variety of environmental medias, including anaerobic sediments, freshwater, and soils, leading to the formation of methylmercury. In addition, more than 10% of inorganic mercury was converted to methylmercury by several methanogens in pure culture systems. Methanogens are crucial to the production of methylmercury. However, there is a lack of systematic summary of mercury methylation in methanogens. In this paper, the role of methanogens in mercury methylation was summarized from the perspectives of in situ environment, laboratory pure culture, and co-culture systems. Based on this summary, the possible mercury methylation pathways of methanogens and future research directions were proposed and discussed. It was suggested that the mercury methylation process may be functionally related to one-carbon metabolism.
-
Key words:
- methanogens /
- methylation /
- methylmercury /
- metabolic pathways.
-

-
表 1 原位环境中与汞甲基化相关的产甲烷菌
Table 1. Mercury methylation-related methanogens in situ habitat
原位环境
In situ habitat位点
Site产甲烷菌
Methanogen文献
Reference沉积物 河口沉积物 圣哈辛托河(美国) 甲烷八叠球菌科Methanosarcinaceae [10] 河流沉积物 橡树岭东部(美国) 甲烷鬃毛菌科Methanosaetaceae
甲烷八叠球菌科Methanosarcinaceae
甲烷球菌科Methanococcaceae[13] 河流沉积物 圣劳伦斯河(加拿大) — [22] 河流沉积物 圣莫里斯河(加拿大) 甲烷绳菌属Methanolinea
Methanoregula*
甲烷螺菌属Methanospirillum[23] 湖泊沉积物 瑞典北部 马赛球菌属Methanomassiliicoccus
Methanoregula*[24] 海洋沉积物 智利、挪威、南极、波罗的海等 马赛球菌属Methanomassiliicoccus
甲烷螺菌属Methanospirillum
甲烷叶菌属Methanolobus
甲烷泡菌属Methanofollis
甲烷胞菌属Methanocella
甲烷粒菌属Methanocorpusculum
Methanoregula*[18] 水体 水生周丛植物
生物膜圣劳伦斯河(加拿大) 甲烷杆菌目Methanobacteriales
甲烷球菌目Methanococcales
甲烷八叠球菌目Methanosarcinales[11] 富营养水体 巢湖、东湖、滇池(中国) — [14] 海水 赤道北部太平洋 马赛球菌属Methanomassiliicoccus [25] 海水 南极 甲烷微菌纲Methanomicrobia [26] 土壤 稻田土壤 万山矿区、铜仁矿区、凤凰矿区
(中国)甲烷螺菌属Methanospirillum
Methanosphaerula*
Methanoregula*
马赛球菌属Methanomassiliicoccus
甲烷叶菌属Methanolobus
甲烷泡菌属Methanofollis
甲烷粒菌属Methanocorpusculum
甲烷胞菌属Methanocella
甲烷球菌纲Methanococci
甲烷杆菌纲Methanobacteria[12, 15, 27] 稻田土壤 阿肯色大学水稻研究和
推广中心(美国)Methanoregula*
甲烷胞菌属Methanocella
甲烷螺菌属Methanospirillum[28] 泥炭地 波的尼亚湾
(瑞典)Methanoregulaceae*
马赛球菌科Methanomassiliicoccaceae
甲烷杆菌科Methanobacteriaceae
Methanoregulaceae*
Methanotrichaceae*[29] 沼泽地土壤 大沼泽地国家森林公园
(美国)甲烷胞菌属Methanoregula
甲烷叶菌属Methanolobus
Methanoregula*[30] 湿地土壤 瑞典北部 甲烷八叠球菌目Methanosarcinales
热源体门Thermoplasmatota[31] 高原永久冻土 青藏高原(中国) 甲烷胞菌属Methanocella
Methanoregula*[32] 永久冻土 博南扎河(美国) 甲烷叶菌属Methanolobus
Methanoregula*
马赛球菌属Methanomassiliicoccus[18] 永久冻土 阿拉斯加州(美国)、瑞典、
芬兰等— [33 − 35] 注:“*”表示部分目、科和属暂无中文译名;“—”表示没有检测该环境中产甲烷菌的具体种类.
Note: “*” indicates that some orders, families, and genera do not have Chinese translated names; “—” indicates that the specific species of methanogens in the environment is not detected.表 2 实验室纯培养中的汞甲基化产甲烷菌
Table 2. Mercury methylators of methanogens in laboratory pure cultures
细菌名称
Strain纲
Class目
Order科
Family编号
Number温度/℃
Temperature底物
Substrate产甲烷途径
Methanogenic
pathway甲基化效率/%
Methylation
efficiency
(MeHg/THg)文献
ReferenceMethanofollis liminatans GKZPZ 甲烷微菌纲 甲烷微菌目 甲烷微菌科 DSM 4140 40 H2+CO2/甲酸、丙醇、丁醇 CO2还原途径 0.19—2.25 [16, 55] Methanocorpusculum bavaricum 甲烷微菌纲 甲烷微菌目 甲烷粒菌科 DSM 4179 37 H2+CO2/甲酸、丙醇、丁醇 CO2还原途径 0.24 [16, 56] Methanospirillum hungatei JF-1 甲烷微菌纲 甲烷微菌目 甲烷螺菌科 DSM 864 30—37 H2+CO2/
甲酸CO2还原途径 43.00—64.20 [16, 50] Methanosphaerula palustris E1-9c 甲烷微菌纲 甲烷微菌目 Methanoregulaceae* DSM 19958 30 H2+CO2/
甲酸CO2还原途径 15.00 [16, 57] Methanolobus tindarius 甲烷微菌纲 甲烷八叠球菌目 甲烷八叠球菌科 DSM 2278 37 甲醇、甲胺 甲基营养途径 0.32—22.10 [9, 16, 47] Methanomethylovorans hollandica 甲烷微菌纲 甲烷八叠球菌目 甲烷八叠球菌科 DSM 15978 34—37 甲醇、甲胺、甲硫醇、二甲基硫醚 甲基营养途径 1.03—3.00 [9, 16, 48] Methanocella paludicola SANAE 甲烷微菌纲 甲烷胞菌目 Methanocellaceae* DSM 17711 37 H2+CO2/
甲酸CO2还原途径 8.63 [16, 58] Methanomassiliicoccus luminyensis B10 热源体纲 马赛球菌目 马赛球菌科 DSM 25720 37 甲醇/H2 甲基营养途径 1.01—53.40 [16, 18, 46] 注:八株菌均属于古菌界、广古菌门;“*”表示部分科暂无中文译名.
Note: All the eight strains belong to the Euryarchaeota of Archaea;“*” indicates that some families do not have Chinese translated names. -
[1] DRISCOLL C T, MASON R P, CHAN H M, et al. Mercury as a global pollutant: Sources, pathways, and effects[J]. Environmental Science & Technology, 2013, 47(10): 4967-4983. [2] 史建波, 阴永光, 江桂斌. 汞的分子转化与长距离传输[M]. 北京: 科学出版社, 2019. SHI J B, YIN Y G, JIANG G B. Molecular transformation and long-distance transport of mercury[M]. Beijing: Science Press, 2019 (in Chinese).
[3] TSZ-KI TSUI M, LIU S N, BRASSO R L, et al. Controls of methylmercury bioaccumulation in forest floor food webs[J]. Environmental Science & Technology, 2019, 53(5): 2434-2440. [4] BOENING D W. Ecological effects, transport, and fate of mercury: A general review[J]. Chemosphere, 2000, 40(12): 1335-1351. doi: 10.1016/S0045-6535(99)00283-0 [5] CHÉTELAT J, ACKERMAN J T, EAGLES-SMITH C A, et al. Methylmercury exposure in wildlife: A review of the ecological and physiological processes affecting contaminant concentrations and their interpretation[J]. Science of the Total Environment, 2020, 711: 135117. doi: 10.1016/j.scitotenv.2019.135117 [6] TANG W L, LIU Y R, GUAN W Y, et al. Understanding mercury methylation in the changing environment: Recent advances in assessing microbial methylators and mercury bioavailability[J]. Science of the Total Environment, 2020, 714: 136827. doi: 10.1016/j.scitotenv.2020.136827 [7] MA M, DU H X, WANG D Y. Mercury methylation by anaerobic microorganisms: A review[J]. Critical Reviews in Environmental Science and Technology, 2019, 49(20): 1893-1936. doi: 10.1080/10643389.2019.1594517 [8] PARKS J M, JOHS A, PODAR M, et al. The genetic basis for bacterial mercury methylation[J]. Science, 2013, 339(6125): 1332-1335. doi: 10.1126/science.1230667 [9] GILMOUR C C, PODAR M, BULLOCK A L, et al. Mercury methylation by novel microorganisms from new environments[J]. Environmental Science & Technology, 2013, 47(20): 11810-11820. [10] WANG Y W, ROTH S, SCHAEFER J K, et al. Production of methylmercury by methanogens in mercury contaminated estuarine sediments[J]. FEMS Microbiology Letters, 2020, 367(23): fnaa196. doi: 10.1093/femsle/fnaa196 [11] HAMELIN S, AMYOT M, BARKAY T, et al. Methanogens: Principal methylators of mercury in lake periphyton[J]. Environmental Science & Technology, 2011, 45(18): 7693-7700. [12] VISHNIVETSKAYA T A, HU H Y, VAN NOSTRAND J D, et al. Microbial community structure with trends in methylation gene diversity and abundance in mercury-contaminated rice paddy soils in Guizhou, China[J]. Environmental Science: Processes & Impacts, 2018, 20(4): 673-685. [13] CHRISTENSEN G A, SOMENAHALLY A C, MOBERLY J G, et al. Carbon amendments alter microbial community structure and net mercury methylation potential in sediments[J]. Applied and Environmental Microbiology, 2018, 84(3): e01049-e01017. [14] LEI P, ZHANG J, ZHU J J, et al. Algal organic matter drives methanogen-mediated methylmercury production in water from eutrophic shallow lakes[J]. Environmental Science & Technology, 2021, 55(15): 10811-10820. [15] LIU Y R, JOHS A, BI L, et al. Unraveling microbial communities associated with methylmercury production in paddy soils[J]. Environmental Science & Technology, 2018, 52(22): 13110-13118. [16] GILMOUR C C, BULLOCK A L, McBURNEY A, et al. Robust mercury methylation across diverse methanogenic Archaea[J]. mBio, 2018, 9(2): e02403-e02417. [17] YU R Q, REINFELDER J R, HINES M E, et al. Mercury methylation by the methanogen Methanospirillum hungatei[J]. Applied and Environmental Microbiology, 2013, 79(20): 6325-6330. doi: 10.1128/AEM.01556-13 [18] PODAR M, GILMOUR C C, BRANDT C C, et al. Global prevalence and distribution of genes and microorganisms involved in mercury methylation[J]. Science Advances, 2015, 1(9): e1500675. doi: 10.1126/sciadv.1500675 [19] YU R Q, BARKAY T. Microbial mercury transformations: Molecules, functions and organisms[J]. Advances in Applied Microbiology, 2022, 118: 31-90. [20] DUAN P F, KHAN S, ALI N, et al. Biotransformation fate and sustainable mitigation of a potentially toxic element of mercury from environmental matrices[J]. Arabian Journal of Chemistry, 2020, 13(9): 6949-6965. doi: 10.1016/j.arabjc.2020.06.041 [21] CHRISTENSEN G A, WYMORE A M, KING A J, et al. Development and validation of broad-range qualitative and clade-specific quantitative molecular probes for assessing mercury methylation in the environment[J]. Applied and Environmental Microbiology, 2016, 82(19): 6068-6078. doi: 10.1128/AEM.01271-16 [22] AVRAMESCU M L, YUMVIHOZE E, HINTELMANN H, et al. Biogeochemical factors influencing net mercury methylation in contaminated freshwater sediments from the St. Lawrence River in Cornwall, Ontario, Canada[J]. Science of the Total Environment, 2011, 409(5): 968-978. doi: 10.1016/j.scitotenv.2010.11.016 [23] MILLERA FERRIZ L, PONTON D E, STORCK V, et al. Role of organic matter and microbial communities in mercury retention and methylation in sediments near Run-of-river hydroelectric dams[J]. Science of the Total Environment, 2021, 774: 145686. doi: 10.1016/j.scitotenv.2021.145686 [24] BRAVO A G, PEURA S, BUCK M, et al. Methanogens and iron-reducing bacteria: The overlooked members of mercury-methylating microbial communities in boreal lakes[J]. Applied and Environmental Microbiology, 2018, 84(23): e01774-e01718. [25] BOWMAN K L, COLLINS R E, AGATHER A M, et al. Distribution of mercury-cycling genes in the Arctic and equatorial Pacific Oceans and their relationship to mercury speciation[J]. Limnology and Oceanography, 2019, 65: S310-S320. [26] SHARMA GHIMIRE P, TRIPATHEE L, ZHANG Q G, et al. Microbial mercury methylation in the cryosphere: Progress and prospects[J]. The Science of the Total Environment, 2019, 697: 134150. doi: 10.1016/j.scitotenv.2019.134150 [27] LIU Y R, YU R Q, ZHENG Y M, et al. Analysis of the microbial community structure by monitoring an Hg methylation gene (hgcA) in paddy soils along an Hg gradient[J]. Applied and Environmental Microbiology, 2014, 80(9): 2874-2879. doi: 10.1128/AEM.04225-13 [28] ROTHENBERG S E, ANDERS M, AJAMI N J, et al. Water management impacts rice methylmercury and the soil microbiome[J]. Science of the Total Environment, 2016, 572: 608-617. doi: 10.1016/j.scitotenv.2016.07.017 [29] WANG B L, HU H Y, BISHOP K, et al. Microbial communities mediating net methylmercury formation along a trophic gradient in a peatland chronosequence[J]. Journal of Hazardous Materials, 2023, 442: 130057. doi: 10.1016/j.jhazmat.2022.130057 [30] SCHAEFER J K, KRONBERG R M, MOREL F M M, et al. Detection of a key Hg methylation gene, hgcA, in wetland soils[J]. Environmental Microbiology Reports, 2014, 6(5): 441-447. doi: 10.1111/1758-2229.12136 [31] XU J Y, LIEM-NGUYEN V, BUCK M, et al. Mercury methylating microbial community structure in boreal wetlands explained by local physicochemical conditions[J]. Frontiers in Environmental Science, 2021, 8: 518662. doi: 10.3389/fenvs.2020.518662 [32] LIU Y R, DONG J X, ZHANG Q G, et al. Longitudinal occurrence of methylmercury in terrestrial ecosystems of the Tibetan Plateau[J]. Environmental Pollution, 2016, 218: 1342-1349. doi: 10.1016/j.envpol.2016.08.093 [33] MACKELPRANG R, WALDROP M P, DeANGELIS K M, et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw[J]. Nature, 2011, 480(7377): 368-371. doi: 10.1038/nature10576 [34] YANG Z M, FANG W, LU X, et al. Warming increases methylmercury production in an Arctic soil[J]. Environmental Pollution, 2016, 214: 504-509. doi: 10.1016/j.envpol.2016.04.069 [35] TARBIER B, HUGELIUS G, KRISTINA SANNEL A B, et al. Permafrost thaw increases methylmercury formation in subarctic fennoscandia[J]. Environmental Science & Technology, 2021, 55(10): 6710-6717. [36] COMPEAU G C, BARTHA R. Sulfate-reducing bacteria: Principal methylators of mercury in anoxic estuarine sediment[J]. Applied and Environmental Microbiology, 1985, 50(2): 498-502. doi: 10.1128/aem.50.2.498-502.1985 [37] FLEMING E J, MACK E E, GREEN P G, et al. Mercury methylation from unexpected sources: Molybdate-inhibited freshwater sediments and an iron-reducing bacterium[J]. Applied and Environmental Microbiology, 2006, 72(1): 457-464. doi: 10.1128/AEM.72.1.457-464.2006 [38] YUAN K, CHEN X, CHEN P, et al. Mercury methylation-related microbes and genes in the sediments of the Pearl River Estuary and the South China Sea[J]. Ecotoxicology and Environmental Safety, 2019, 185: 109722. doi: 10.1016/j.ecoenv.2019.109722 [39] 周心劝. 稻田土壤中微生物群落对甲基汞积累的影响[D]. 重庆: 西南大学, 2019. ZHOU X Q. Effects of microbial communities on the accumulation of methylmercury in paddy soils[D]. Chongqing: Southwest University, 2019 (in Chinese).
[40] ZHAO L, MENG B, FENG X B. Mercury methylation in rice paddy and accumulation in rice plant: A review[J]. Ecotoxicology and Environmental Safety, 2020, 195: 110462. doi: 10.1016/j.ecoenv.2020.110462 [41] 高润霞, 罗文倩, 胡海艳, 等. 稻田土壤中汞的微生物甲基化研究进展[J]. 宁夏农林科技, 2020, 61(1): 46-49. GAO R X, LUO W Q, HU H Y, et al. Research progress of microbial methylation of mercury in paddy soil[J]. Ningxia Journal of Agriculture and Forestry Science and Technology, 2020, 61(1): 46-49 (in Chinese).
[42] CLARKE R T, HUNGATE R E. Culture of the rumen holotrich ciliate Dasytricha ruminantium schuberg[J]. Applied Microbiology, 1966, 14(3): 340-345. doi: 10.1128/am.14.3.340-345.1966 [43] SMITH P H, HUNGATE R E. Isolation and characterization of Methanobacterium ruminantium n. sp[J]. Journal of Bacteriology, 1958, 75(6): 713-718. doi: 10.1128/jb.75.6.713-718.1958 [44] BRYANT M P, WOLIN E A, WOLIN M J, et al. Methanobacillus omelianskii, a symbiotic association of two species of bacteria[J]. Archiv Fur Mikrobiologie, 1967, 59(1): 20-31. [45] WOOD J M, KENNEDY F S, ROSEN C G. Synthesis of methyl-mercury compounds by extracts of a methanogenic bacterium[J]. Nature, 1968, 220(5163): 173-174. doi: 10.1038/220173a0 [46] DRIDI B, FARDEAU M L, OLLIVIER B, et al. Methanomassiliicoccus luminyensis Gen nov sp nov , a methanogenic archaeon isolated from human faeces[J]. International Journal of Systematic and Evolutionary Microbiology, 2012, 62(Pt 8): 1902-1907. [47] KÖNIG H, STETTER K O. Isolation and characterization of Methanolobus tindarius sp nov, a coccoid methanogen growing only on methanol and methylamines[J]. Zentralblatt Für Bakteriologie Mikrobiologie Und Hygiene: I. Abt. Originale C: Allgemeine, Angewandte Und Ökologische Mikrobiologie, 1982, 3(4): 478-490. [48] LOMANS B P, MAAS R, LUDERER R, et al. Isolation and characterization of Methanomethylovorans hollandica Gen nov sp nov , isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol[J]. Applied and Environmental Microbiology, 1999, 65(8): 3641-3650. [49] LIU Y C, WHITMAN W B. Metabolic, phylogenetic, and ecological diversity of the methanogenic Archaea[J]. Annals of the New York Academy of Sciences, 2008, 1125(1): 171-189. doi: 10.1196/annals.1419.019 [50] FERRY J G, SMITH P H, WOLFE R S. Methanospirillum, a new genus of methanogenic bacteria, and characterization of Methanospirillum hungatii sp. nov[J]. International Journal of Systematic Bacteriology, 1974, 24(4): 465-469. doi: 10.1099/00207713-24-4-465 [51] BENOIT J M, GILMOUR C C, MASON R P, et al. Sulfide controls on mercury speciation and bioavailability to methylating bacteria in sediment pore waters[J]. Environmental Science & Technology, 1999, 33(6): 951-957. [52] BENOIT J M, GILMOUR C C, MASON R P. Aspects of bioavailability of mercury for methylation in pure cultures of Desulfobulbus propionicus (1pr3)[J]. Applied and Environmental Microbiology, 2001, 67(1): 51-58. doi: 10.1128/AEM.67.1.51-58.2001 [53] OREM W, GILMOUR C, AXELRAD D, et al. Sulfur in the south Florida ecosystem: Distribution, sources, biogeochemistry, impacts, and management for restoration[J]. Critical Reviews in Environmental Science and Technology, 2011, 41(sup1): 249-288. doi: 10.1080/10643389.2010.531201 [54] BENOIT J M, GILMOUR C C, HEYES A, et al. Geochemical and biological controls over methylmercury production and degradation in aquatic ecosystems[M]//ACS Symposium Series. Washington, DC: American Chemical Society, 2002: 262-297. [55] ZELLNER G, SLEYTR U B, MESSNER P, et al. Methanogenium liminatans spec nov , a new coccoid, mesophilic methanogen able to oxidize secondary alcohols[J]. Archives of Microbiology, 1990, 153(3): 287-293. [56] ZELLNER G, STACKEBRANDT E, MESSNER P, et al. Methanocorpusculaceae fam. nov, represented by Methanocorpusculum parvum, Methanocorpusculum sinense spec nov and Methanocorpusculum bavaricum spec. nov[J]. Archives of Microbiology, 1989, 151(5): 381-390. doi: 10.1007/BF00416595 [57] CADILLO-QUIROZ H, YAVITT J B, ZINDER S H. Methanosphaerula palustris gen. nov sp nov , a hydrogenotrophic methanogen isolated from a minerotrophic fen peatland[J]. International Journal of Systematic and Evolutionary Microbiology, 2009, 59(Pt 5): 928-935. [58] SAKAI S, IMACHI H, HANADA S, et al. Methanocella paludicola gen. nov sp nov , a methane-producing archaeon, the first isolate of the lineage 'Rice Cluster I', and proposal of the new archaeal order Methanocellales ord. nov[J]. International Journal of Systematic and Evolutionary Microbiology, 2008, 58(Pt 4): 929-936. [59] YU R Q, REINFELDER J R, HINES M E, et al. Syntrophic pathways for microbial mercury methylation[J]. The ISME Journal, 2018, 12(7): 1826-1835. doi: 10.1038/s41396-018-0106-0 [60] PAK K, BARTHA R. Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens[J]. Applied and Environmental Microbiology, 1998, 64(6): 1987-1990. doi: 10.1128/AEM.64.6.1987-1990.1998 [61] ABRAM J W, NEDWELL D B. Inhibition of methanogenesis by sulphate reducing bacteria competing for transferred hydrogen[J]. Archives of Microbiology, 1978, 117(1): 89-92. doi: 10.1007/BF00689356 [62] BRYANT M P, CAMPBELL L L, REDDY C A, et al. Growth of desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria[J]. Applied and Environmental Microbiology, 1977, 33(5): 1162-1169. doi: 10.1128/aem.33.5.1162-1169.1977 [63] PHELPS T J, CONRAD R, ZEIKUS J G. Sulfate-dependent interspecies H2 transfer between Methanosarcina barkeri and Desulfovibrio vulgaris during coculture metabolism of acetate or methanol[J]. Applied and Environmental Microbiology, 1985, 50(3): 589-594. doi: 10.1128/aem.50.3.589-594.1985 [64] KRIVORUCHKO A, ZHANG Y M, SIEWERS V, et al. Microbial acetyl-CoA metabolism and metabolic engineering[J]. Metabolic Engineering, 2015, 28: 28-42. doi: 10.1016/j.ymben.2014.11.009 [65] NIKOLAU B J, OLIVER D J, SCHNABLE P S, et al. Molecular biology of acetyl-CoA metabolism[J]. Biochemical Society Transactions, 2000, 28(6): 591-593. doi: 10.1042/bst0280591 [66] SCHÖNE C, POEHLEIN A, JEHMLICH N, et al. Deconstructing Methanosarcina acetivorans into an acetogenic archaeon[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(2): e2113853119. [67] KIM Y T, JUNG J H, STEWART L C, et al. Complete genome sequence of the hyperthermophilic methanogen Methanocaldococcus bathoardescens JH146T isolated from the basalt subseafloor[J]. Marine Genomics, 2015, 24: 229-230. doi: 10.1016/j.margen.2015.06.002 [68] LINDAHL P A, CHANG B. The evolution of acetyl-CoA synthase[J]. Origins of Life and Evolution of the Biosphere, 2001, 31(4): 403-434. [69] MARTIN W F. Older than genes: The acetyl CoA pathway and origins[J]. Frontiers in Microbiology, 2020, 11: 817. doi: 10.3389/fmicb.2020.00817 [70] CHOI S C, CHASE T, BARTHA R. Metabolic pathways leading to mercury methylation in Desulfovibrio desulfuricans LS[J]. Applied and Environmental Microbiology, 1994, 60(11): 4072-4077. doi: 10.1128/aem.60.11.4072-4077.1994 [71] BERMAN M, CHASE T, BARTHA R. Carbon flow in mercury biomethylation by Desulfovibrio desulfuricans[J]. Applied and Environmental Microbiology, 1990, 56(1): 298-300. doi: 10.1128/aem.56.1.298-300.1990 [72] FINKELSTEIN J D. Methionine metabolism in mammals[J]. The Journal of Nutritional Biochemistry, 1990, 1(5): 228-237. doi: 10.1016/0955-2863(90)90070-2 [73] DUCKER G S, RABINOWITZ J D. One-carbon metabolism in health and disease[J]. Cell Metabolism, 2017, 25(1): 27-42. doi: 10.1016/j.cmet.2016.08.009 [74] RODIONOV D A, VITRESCHAK A G, MIRONOV A A, et al. Comparative genomics of the methionine metabolism in Gram-positive bacteria: A variety of regulatory systems[J]. Nucleic Acids Research, 2004, 32(11): 3340-3353. doi: 10.1093/nar/gkh659 [75] BANERJEE R V, MATTHEWS R G. Cobalamin-dependent methionine synthase[J]. The FASEB Journal, 1990, 4(5): 1450-1459. doi: 10.1096/fasebj.4.5.2407589 [76] FROESE D S, FOWLER B, BAUMGARTNER M R. Vitamin B12, folate, and the methionine remethylation cycle—Biochemistry, pathways, and regulation[J]. Journal of Inherited Metabolic Disease, 2019, 42(4): 673-685. doi: 10.1002/jimd.12009 [77] PANAYIOTIDIS M I, STABLER S P, AHMAD A, et al. Activation of a novel isoform of methionine adenosyl transferase 2A and increased S-adenosylmethionine turnover in lung epithelial cells exposed to hyperoxia[J]. Free Radical Biology and Medicine, 2006, 40(2): 348-358. doi: 10.1016/j.freeradbiomed.2005.09.004 [78] WANG P P, BAO P, SUN G X. Identification and catalytic residues of the arsenite methyltransferase from a sulfate-reducing bacterium, Clostridium sp . BXM[J]. FEMS Microbiology Letters, 2015, 362(1): 1-8. [79] LANDNER L. Biochemical Model for the Biological Methylation of Mercury suggested from Methylation Studies in vivo with Neurospora crassa[J]. Nature, 1971, 230(5294): 452-454. doi: 10.1038/230452a0 [80] EKSTROM E B, MOREL F M M, BENOIT J M. Mercury methylation independent of the acetyl-coenzyme A pathway in sulfate-reducing bacteria[J]. Applied and Environmental Microbiology, 2003, 69(9): 5414-5422. doi: 10.1128/AEM.69.9.5414-5422.2003 [81] EKSTROM E B, MOREL F M M. Cobalt limitation of growth and mercury methylation in sulfate-reducing bacteria[J]. Environmental Science & Technology, 2008, 42(1): 93-99. [82] REISINGER K, STOEPPLER M, NÜRNBERG H W. Methylcarbenium ion transfer from S‐adenosylmethionine to inorganic mercury—One of the possible biological pathways to methylmercury?[J]. Toxicological & Environmental Chemistry, 1984, 8(1): 45-54. [83] OLTEANU H, BANERJEE R. Human methionine synthase reductase, a soluble P-450 reductase-like dual flavoprotein, is sufficient for NADPH-dependent methionine synthase activation[J]. Journal of Biological Chemistry, 2001, 276(38): 35558-35563. doi: 10.1074/jbc.M103707200 [84] WILSON A, LECLERC D, ROSENBLATT D S, et al. Molecular basis for methionine synthase reductase deficiency in patients belonging to the cblE complementation group of disorders in folate/cobalamin metabolism[J]. Human Molecular Genetics, 1999, 8(11): 2009-2016. doi: 10.1093/hmg/8.11.2009 [85] ZHOU J, RICCARDI D, BESTE A, et al. Mercury methylation by HgcA: Theory supports carbanion transfer to Hg(II)[J]. Inorganic Chemistry, 2014, 53(2): 772-777. doi: 10.1021/ic401992y [86] WOOD J M. Biological cycles for toxic elements in the environment[J]. Science, 1974, 183(4129): 1049-1052. doi: 10.1126/science.183.4129.1049 [87] LI J J, SUN C X, CAI W W, et al. Insights into S-adenosyl-l-methionine (SAM)-dependent methyltransferase related diseases and genetic polymorphisms[J]. Mutation Research. Reviews in Mutation Research, 2021, 788: 108396. doi: 10.1016/j.mrrev.2021.108396 [88] 方晓瑜, 李家宝, 芮俊鹏, 等. 产甲烷生化代谢途径研究进展[J]. 应用与环境生物学报, 2015, 21(1): 1-9. FANG X Y, LI J B, RUI J P, et al. Research progress in biochemical pathways of methanogenesis[J]. Chinese Journal of Applied and Environmental Biology, 2015, 21(1): 1-9 (in Chinese).
[89] BOTT M H, EIKMANNS B, THAUER R K. Defective formation and/or utilization of carbon monoxide in H2/CO2 fermenting methanogens dependent on acetate as carbon source[J]. Archives of Microbiology, 1985, 143(3): 266-269. doi: 10.1007/BF00411248 [90] THAUER R K. Biochemistry of methanogenesis: A tribute to marjory stephenson: 1998 marjory stephenson prize lecture[J]. Microbiology, 1998, 144 ( Pt 9): 2377-2406. -



 下载:
下载: