-
聚丙烯酰胺(PAM)是一种线性水溶性高分子化合物,作为絮凝剂广泛用于选矿废水的处理[1]。PAM按带电性质不同分为非离子型聚丙烯酰胺(NPAM)、阳离子型聚丙烯酰胺(CPAM)、阴离子型聚丙烯酰胺(HPAM)和两性离子型聚丙烯酰胺(ZPAM)[2]。NPAM与黏土之间的吸附通过聚合物C=O基团与黏土可交换阳离子之间离子耦合和氢键作用而增强。含有大量N+活性基团的CPAM很容易吸附在带负电的黏土上,而HPAM通过多价阳离子在其COO−与黏土表面实现架桥[3]。ZPAM兼有CPAM和HPAM的特点,适应范围更广。在细煤的反浮选过程中,发现阴离子度50%的HPAM是最有效的细煤絮凝剂[4]。HPAM通过吸附架桥作用,即多价阳离子优先在细煤颗粒与HPAM上COO−之间充当桥梁,而HPAM的高分子链与细煤形成疏水键,最终形成微米级团聚体以提高HPAM对尾煤废水的处理能力。HPAM的大量使用会在选煤循环水中累积,从而降低煤泥对浮选药剂的吸附能力,降低浮选效果[5]。另外,含HPAM的选煤废水外排会造成环境污染[6]。因此,急需研发对煤泥水中HPAM脱除的有效方法。
微生物降解是脱除HPAM的有效方法,具有环境友好、条件温和、无二次污染等优点[7]。Bao等[8]从聚合物驱采出水中分离出两种HPAM降解细菌,即Bacillus cereus 和Bacillus sp.,同时假设了好氧细菌的降解机理。Wen等[9]从油田的活性淤泥和土壤里分别分离出Bacillus cereus 和Bacillus flexu,实验发现它们可以从HPAM主链上攻击酰胺基团。HPAM侧链上的酰胺基团很容易受酰胺酶进攻并引发水解成羧基[10-11]。此外,通过HPLC分析发现不存在单体丙烯酰胺的峰,说明在HPAM生物降解过程中没有丙烯酰胺单体的产生[8-9]。聚丙烯酸酯(PAA)是HPAM经脱氨基后残留的高分子碳骨架。PAA具有较强的生物抗性,但微生物仍能利用PAA作为营养物质-碳源[12]。生物降解的本质是酶促反应,降解关键在于获得高活性的降解酶。但通过实验无法得到酰胺酶与底物相互作用的细节,而且从环境中筛选得到的酶活性一般较低,导致降解效果欠佳,费时费力[13]。天然的酶可以通过定点突变技术来进行理性设计,目的是提高酶的活性。底物与酰胺酶的活性中心的微观作用为定点突变提供了理论依据,从而设计出更高活性的酶。此外,还有助于对酶促降解机理进行深入理解。
分子对接是一种生物信息学技术,可以搜索到受体蛋白与配体最优的结合构象,一般用于药物设计,但现在也常用于发现环境中有机污染物与酶活性位点的最佳结合模式[14]。酰胺酶的三维晶体结构早期通过实验已经解析出来,这为进一步在分子水平上研究酰胺酶与底物相互作用提供了可能性[15]。目前对HPAM的生物降解仍停留在实验初级阶段,通过实验只能检测到HPAM降解过程中酰胺酶的存在[16]。但从分子水平上研究其与底物的相互作用还未见报道。
本文采用Discovery Studio (DS) 2020软件的CDOCKER半柔性对接程序进行红球菌酰胺酶与HPAM或PAA的分子对接研究,然后基于亲和力的虚拟突变对最大亲和力的酶-底物复合物进行丙氨酸(ALA)扫描。该研究的结果用于理解生物降解及设计酶的高活性突变体,以期彻底降解煤泥水中残留的HPAM。
-
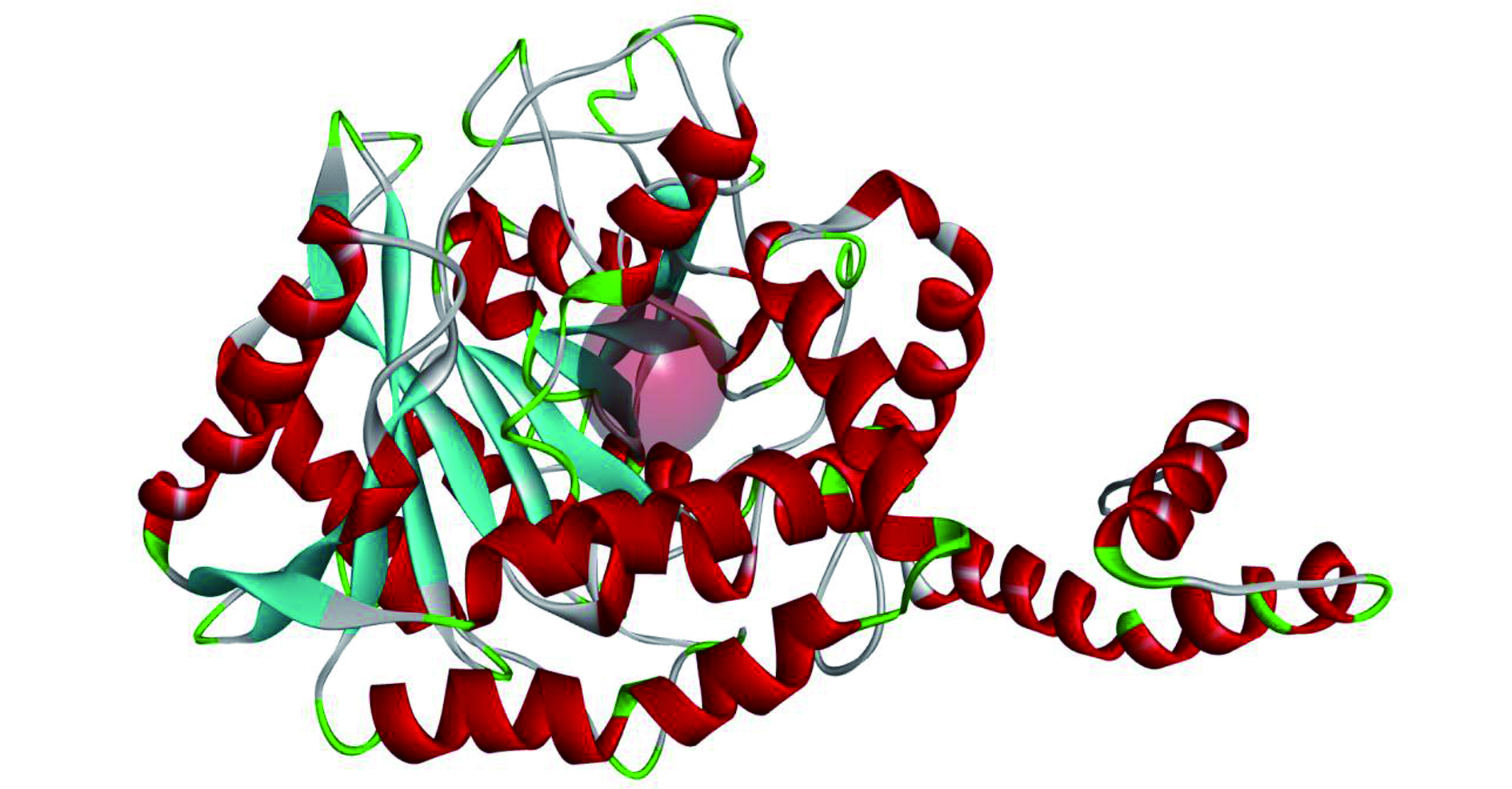
酰胺酶的活性已经在很多菌属中被识别和鉴定,包括红球菌属、芽孢杆菌属、分枝杆菌属、产碱杆菌属、假单胞菌属等[17-18]。酰胺酶的三维结构文件从RCSB网站下载得到,对应的PDB编号为3A1I,分辨率为0.232 nm,A链,该酶[19]来源:Rhodococcus sp. N-771,简称为Rh Amidase。采用DS 2020软件的Prepare Protein工具对酰胺酶进行预处理,去水加氢、修补缺失的氨基酸残基。图1就是预处理的Rh Amidase,其中透明红色球体代表用于对接活性位点的位置,坐标(X, -20.835; Y, 6.34554; Z, -5.67827; Radius, 5)。
-
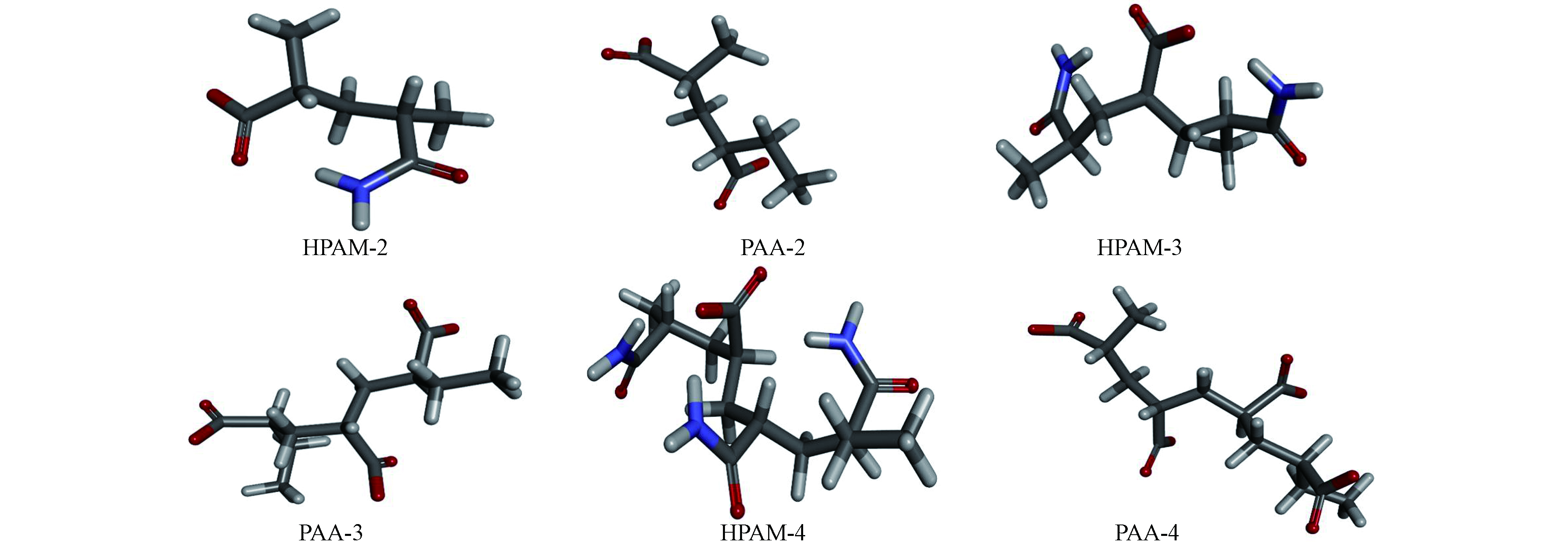
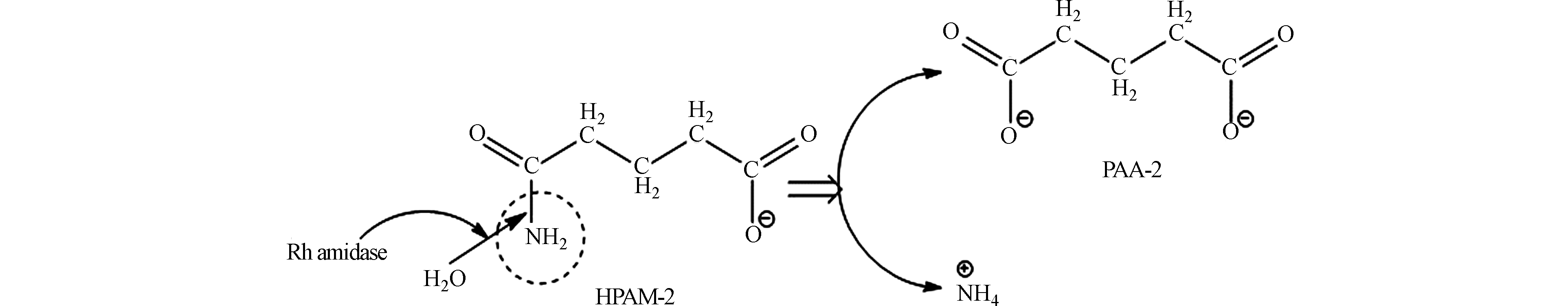
酶催化降解底物主要涉及官能团之间发生的反应,本文仅考虑分子主链上的高反应活性官能团之间的相互作用。HPAM也称为部分水解的PAM,在弱碱性环境下其羧基侧链带负电。PAA实际上是HPAM的脱氨产物。它们的结构模型采用Materials Studio 2017软件的Build polymer工具绘制,两端采用甲基进行封端,同样的处理见文献[20]。最后采用CHARMm力场对其构型进行优化。图2显示了几种代表不同分子链长度的HPAM和PAA模型物优化构型。
-
采用DS 2020软件的CDOCKER半柔性对接程序分别进行Rh Amidase与HPAM或PAA对接模拟实验[21]。CDOCKER对接程序基于高温动力学产生配体构象集,同时采用模拟退火方法对各个配体分子柔性构象依次在受体蛋白活性位点处进行优化。对接结果默认参数设置为保留最优配体的前10种构象,用-CDOCKER_ Energy score值大小进行排序,选择打分最高的配体构象进行分析。
-
底物与活性口袋处氨基酸残基的非键相互作用范围一般为0.5 nm。为了进一步确定亲和力最大的酶-底物复合物中的关键氨基酸残基,因此采用Calculate Mutation Energy (Binding)程序对配体周围0.5 nm范围内的氨基酸残基进行ALA扫描,选择突变后突变能最大的氨基酸残基作为关键氨基酸残基,而突变能最小的氨基酸残基可以作为理论设计高活性酶蛋白的突变点。
-
采用CDOCKER对接程序将原始配体苯甲酰胺对接到Rh Amidase的活性中心,对接结果默认参数设置为保留最优的配体前10种构象。对获得的10种配体构象分别与晶体结构中的原始配体进行对比,可以发现-CDOCKER_ Energy score值最高的构象与天然晶体结构中的苯甲酰胺具有较好的叠合效果,如图3所示,其中绿色代表晶体结构中原始配体的位置。它们之间均方根偏差值(RMSD)为0.04033 nm,RMSD偏差小于0.2 nm,说明该对接程序的准确度是可靠的[22]。
-
CDOCKER主要计算两种能量打分来评估对接结果的好坏[23]。即-CDOCKER_ Energy score是酶和底物总的能量打分,其值越大越说明该体系的亲和力越大,越稳定;-CDOCKER_ Interaction_ Energy score值越大越说明对接过程中相互作用越小,酶与底物结合的越好。从对接结果(表1)中可以发现,Rh Amidase-HPAM-2拥有最高的-CDOCKER_ Energy score值,说明该酶对HPAM-2有最高的亲和力[24];Rh Amidase-PAA-2拥有最高的-CDOCKER_ Interaction_ Energy score值,说明Rh Amidase与PAA-2相互作用最小,它们结合最好。但他们的亲和力及稳定性(36.085 kcal·mol−1)略低于Rh Amidase-HPAM-2(37.050 kcal·mol−1),说明HPAM-2经Rh Amidase水解后的PAA-2可以作为微生物的碳源,比HPAM-2更容易脱离酶的活性中心。无论是基于哪种能量打分的标准,Rh Amidase对HPAM和PAA的打分皆随其碳链的延长而迅速下降,说明Rh Amidase倾向于降解短链的低分子量的寡聚物。
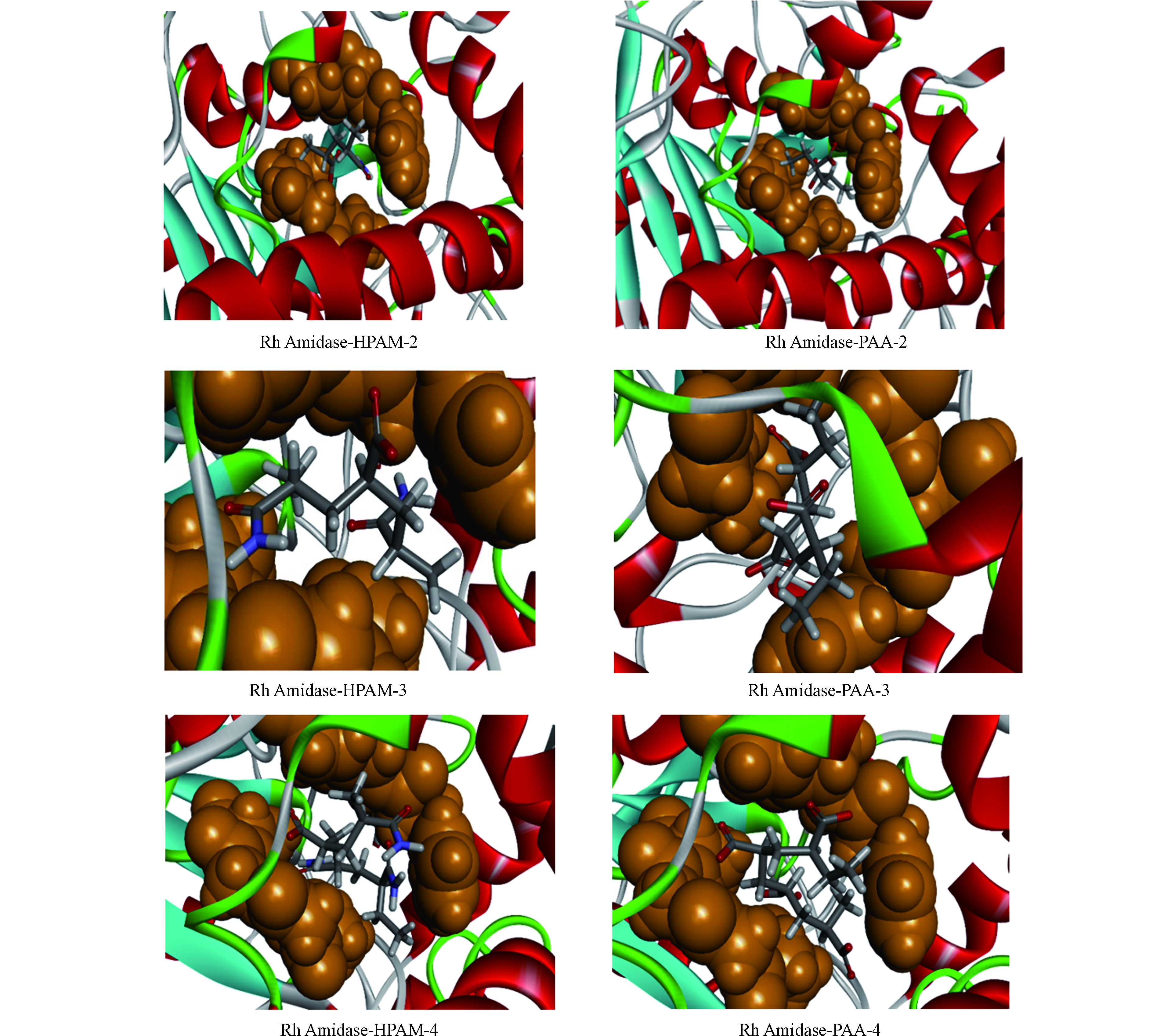
图4显示了HPAM和PAA与Rh Amidase最佳的结合构象,橙色代表该酶的活性位点。HPAM和PAA皆随聚合度的增加而向活性位点外伸展,说明高分子量的聚合物很难被Rh Amidase降解。同时也发现,HPAM-2和PAA-2都完全被包裹在该酶的活性位点上,这可以解释Rh Amidase-HPAM-2和Rh Amidase-PAA-2皆有相应较高的能量打分值。
-
底物与活性口袋附近氨基酸残基的具体作用细节对酶催化底物是至关重要的。通过DS 2020将能量打分较高的Rh Amidase-HPAM-2和Rh Amidase-PAA-2进一步可视化,显示其相互作用关系。从图5可以观察到,HPAM-2和PAA-2的羧基阴离子均与SER171、GLY193、ALA195和GLN192等氨基酸残基形成4个氢键(绿色)。HPAM-2的氨基与CYS145形成一个氢键,而该氨基水解形成氧负离子后,羰基与SER147形成一个碳氢键(浅蓝色)。HPAM-2高分子链两端与ALA195、ILE198、ILE450、TRP328、PHE146和LEU447等氨基酸残基形成7个疏水键(粉色);PAA-2高分子链两端与PHE146、ILE223、ILE198和ILE450等氨基酸残基形成4个疏水键。通过对比发现,Rh Amidase-HPAM-2比Rh Amidase-PAA-2多了3个疏水相互作用。这说明Rh Amidase-HPAM-2比Rh AmidasePAA-2更稳定主要是由于疏水相互作用。
目前实验研究主要集中在好氧细菌降解HPAM上,实验结果表明[8-10],HPAM的酰胺侧基被酰胺酶水解成羧基,NH4+被释放到溶液中作为微生物利用的唯一氮源。分子对接的结果表明,HPAM-2的酰胺侧链经Rh Amidase水解成COO−后,PAA-2与酶的疏水相互作用减弱,可以较好地脱离该酶的催化活性中心。文献[11]报道的细菌更容易利用HPAM的酰胺侧链为氮源,该试验结果与上述模拟结果相互印证。
-
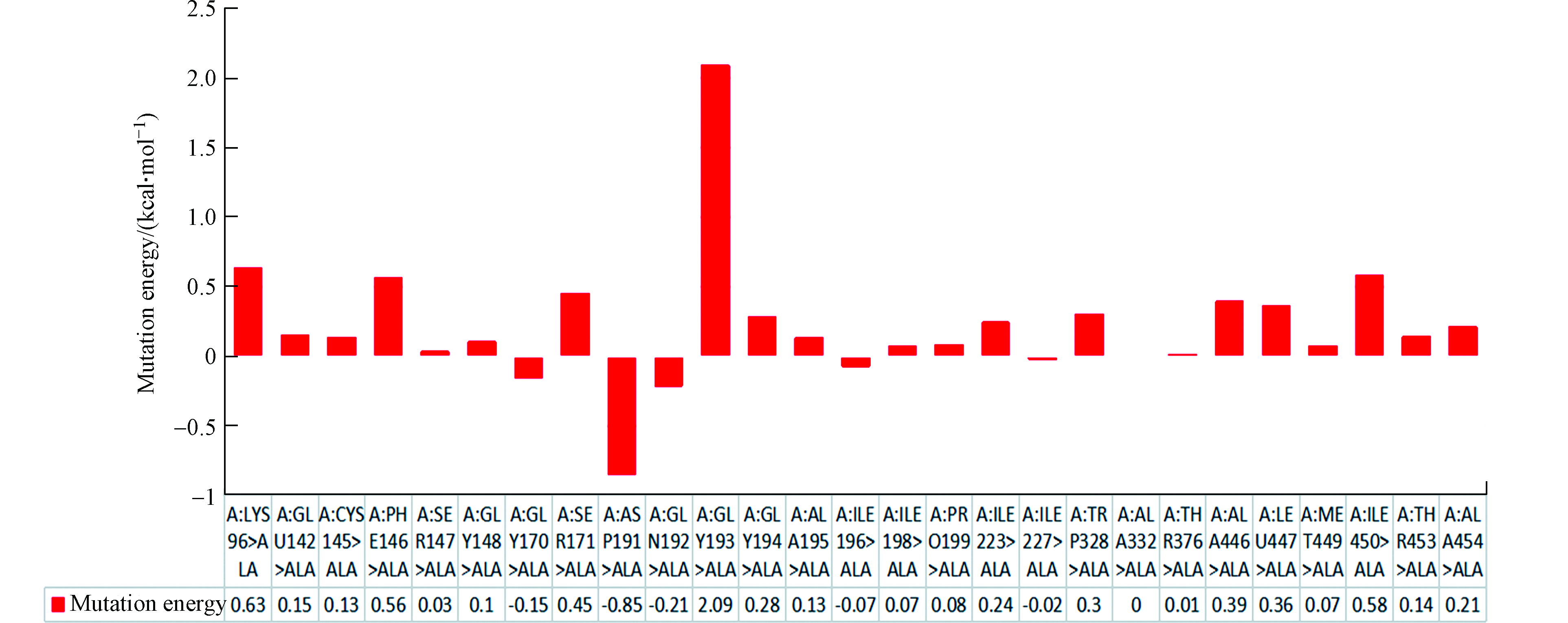
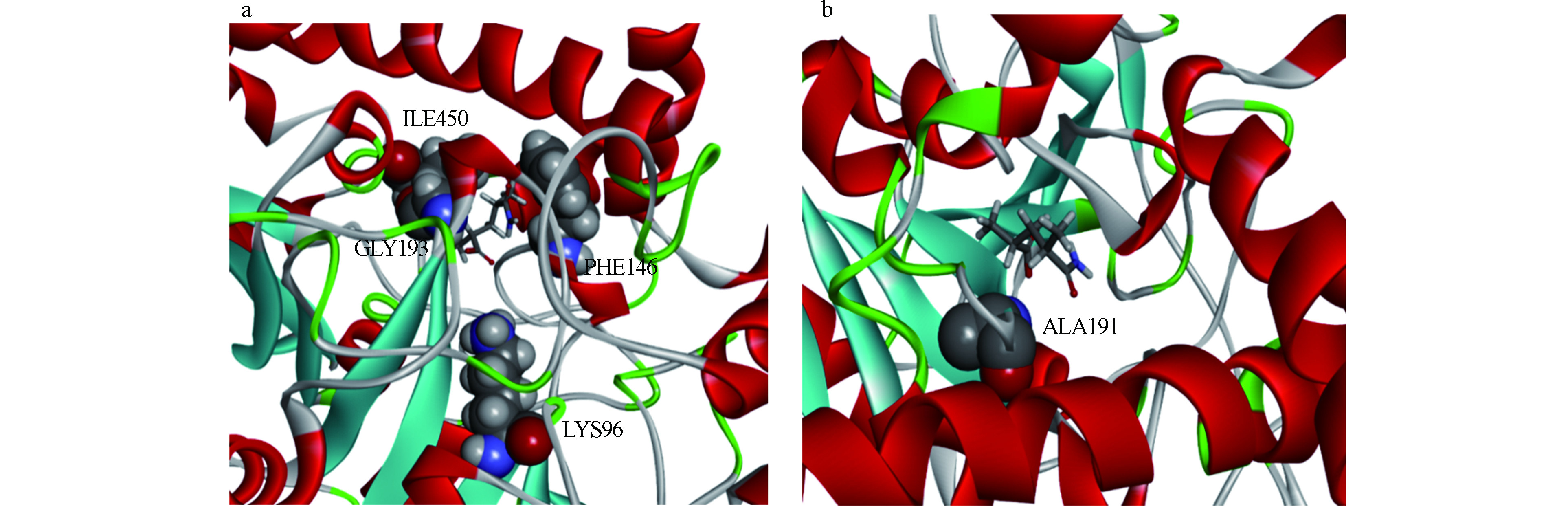
突变能是酶与底物复合物突变后能量的变化。突变能作为唯一的标准用于评估点突变对Rh Amidase-HPAM-2亲和力的影响。图6显示了该复合物ALA扫描所有的突变能,而表2只列出了5个最高的突变能和5个最低的突变能。从Rh Amidase-HPAM-2 ALA扫描的结果可以发现, PHE146、ILE450、LYS96和GLY193突变为ALA,突变能均大于0.50 kcal·mol−1,效果为不稳定,表明这种突变会使酶与底物的亲和力下降,由此推测出这4种氨基酸残基是Rh Amidase与HPAM-2相互作用的关键氨基酸残基。其中GLY193ALA突变能(2.09 kcal·mol−1)最大,说明GLY193对Rh Amidase与HPAM-2亲和力的影响最大,即GLY193与HPAM-2形成的1个氢键对Rh Amidase-HPAM-2亲和力影响最大。图7(a)显示了Rh Amidase-HPAM-2的关键氨基酸残基,其中关键氨基酸残基通过CPK显示。当ASP191突变为ALA时,突变能为-0.85 kcal·mol−1,效果为稳定,表明该突变可以增强Rh Amidase对HPAM-2的亲和力,即提高Rh Amidase对HPAM-2的酶活力。图7(b)显示了Rh Amidase-HPAM-2的ASP191ALA突变体,其中ASP191突变为ALA通过CPK显示。
-
根据上述模拟得到的结果,Rh Amidase介导的HPAM生物降解机理的假设被提出来。Rh Amidase倾向于水解低分子量HPAM的酰胺侧链,由于Rh Amidase-HPAM-2亲和力最大、最稳定,因此选择HPAM-2的酶解为例,尝试探究HPAM的生物降解机理。Rh Amidase攻击HPAM-2的酰胺侧链,将HPAM-2水解成PAA-2和NH4+,其中NH4+作为微生物生长的氮源,而PAA-2作为水解产物,供微生物生长的碳源,如图8所示。
-
首先通过比较Rh Amidase晶体结构中原始配体与对接结果之间的RMSD值,验证了软件的可靠性。其次通过分子对接获得了Rh Amidase与底物最优的结合构象,从中选择亲和力较大的Rh Amidase-HPAM-2和Rh Amidase-PAA-2进行相互作用细节分析。最后对亲和力最大、最稳定的Rh Amidase-HPAM-2中配体周围0.5 nm范围内残基进行ALA扫描。
(1)通过亲和力分析表明,Rh Amidase-HPAM-2拥有最高的亲和力,最稳定,而Rh Amidase-PAA-2在对接过程中相互作用最小、结合最好。Rh Amidase对HPAM和PAA的亲和力皆随其碳链延长而迅速减弱,说明该酶更容易降解短链的聚合物。
(2)通过相互作用细节分析可以发现,Rh Amidase-HPAM-2比Rh Amidase-PAA-2多3个疏水键,可以推测出疏水相互作用是Rh Amidase-HPAM-2亲和力最大、最稳定的主要原因。
(3)根据ALA扫描得知,PHE146、ILE450、LYS96和GLY193等氨基酸残基是Rh Amidase降解HPAM-2的关键氨基酸残基。其中 GLY193对Rh Amidase-HPAM-2的亲和力影响最大,而且该酶的低活性可以通过ASP191突变为ALA进行改善。
红球菌酰胺酶降解阴离子型聚丙烯酰胺的亲和力分析
Affinity analysis of anionic polyacrylamide degraded by amidase from Rhodococcus sp.N-771
-
摘要: 为探究酰胺酶降解阴离子型聚丙烯酰胺(HPAM)的微观机理,采用分子对接分别模拟了阴离子型聚丙烯酰胺(HPAM)或聚丙烯酸酯(PAA)结构模型与Rhodococcus sp. N-771酰胺酶(Rh Amidase)的结合,根据-CDOCKER ˍ Energy score值最高的原则,对获得最佳结合构象进行分析。基于亲和力虚拟突变进行丙氨酸(ALA)扫描。亲和力分析表明,Rh Amidase对HPAM-2的亲和力最高、最稳定,而Rh Amidase于PAA-2相互作用最小、结合最好。同时,该酶更倾向于降解短链的聚合物。相互作用分析表明,疏水相互作用是Rh Amidase-HPAM-2比Rh Amidase-PAA-2更稳定的主要原因。通过ALA扫描进一步得知,PHE146、ILE450、LYS96和GLY193是Rh Amidase降解HPAM-2的关键氨基酸残基。其中 GLY193与HPAM-2形成的1个氢键对Rh Amidase-HPAM-2的亲和力影响最大。突变体ASP191ALA可以提高Rh Amidase对HPAM-2的酶活性,这些数据可为设计更高活性的Rh Amidase突变体提供理论指导。
-
关键词:
- 酰胺酶 /
- 阴离子型聚丙烯酰胺(HPAM) /
- 分子对接 /
- 丙氨酸(ALA)扫描
Abstract: In order to explore the microscopic degradation mechanism of anionic polyacrylamide (HPAM) by amidase, molecular docking was performed to investigate the binding of anionic polyacrylamide (HPAM) or polyacrylate (PAA) structural model with amidase from Rhodococcus sp. N-771 (Rh Amidase), respectively. The optimal conformations were obtained according to the highest principle of -CDOCKER_ Energy score and analyzed. Then, alanine (ALA) scanning was performed by virtual mutation based on affinity. The analysis of affinity indicated that Rh Amidase-HPAM-2 acquired the highest affinity and most stable, while Rh Amidase had the least interaction and the optimal binding with PAA-2. Meanwhile, this enzyme was more likely to degrade polymers with short chains. The interaction analysis showed that the hydrophobic interaction was the main reason why Rh Amidase-HPAM-2 was more stable than Rh Amidase-PAA-2. It was further confirmed by ALA scanning that the PHE146、ILE450、LYS96 and GLY193 were the key amino acid residues of Rh Amidase degrading HPAM-2. Among them, a hydrogen bond formed by GLY193 with HPAM-2 had the greatest effect on the affinity of Rh Amidase-HPAM-2. The mutant ASP191ALA could improve the enzyme activity of Rh Amidase against HPAM-2. These data provides theoretical guidance for designing Rh Amidase mutants with higher activity.-
Key words:
- amidase /
- anionic polyacrylamide (HPAM) /
- molecular docking /
- alanine (ALA) scanning
-

-
表 1 通过CDOCKER工具获得的对接结果
Table 1. Docking results obtained from CDOCKER protocol
酶与底物复合物
Enzyme-substrate complex负的总能量打分/(kcal·mol−1)
-CDOCKER ˍ Energy score负的相互作用能量打分/(kcal·mol−1)
-CDOCKER ˍ Interaction ˍ Energy scoreRh Amidase-HPAM-2 37.050 38.903 Rh Amidase-HPAM-3 8.479 18.776 Rh Amidase-HPAM-4 −141.214 −25.364 Rh Amidase-PAA-2 36.085 43.735 Rh Amidase-PAA-3 −31.644 14.582 Rh Amidase-PAA-4 −120.304 −32.564 表 2 Rh Amidase-HPAM-2 ALA扫描的结果
Table 2. Results of Rh Amidase-HPAM-2 ALA scanning
突变
Mutation突变能/(kcal·mol−1)
Mutation Energy突变效果
Effect of MutationASP191ALA −0.85 稳定 GLN192ALA −0.21 中性 GLY170ALA −0.15 中性 ILE196ALA −0.07 中性 ILE227ALA −0.02 中性 SER171ALA 0.45 中性 PHE146ALA 0.56 不稳定 ILE450ALA 0.58 不稳定 LYS96ALA 0.63 不稳定 GLY193ALA 2.09 不稳定 -
[1] REN B, MIN F F, LIU L Y, et al. Adsorption of different PAM structural units on kaolinite (0 0 1) surface: Density functional theory study [J]. Applied Surface Science, 2020, 504: 144324. doi: 10.1016/j.apsusc.2019.144324 [2] 包木太, 彭杰, 陈庆国. 微生物对聚丙烯酰胺降解作用的研究进展 [J]. 化工进展, 2011, 30(9): 2080-2086. doi: 10.16085/j.issn.1000-6613.2011.09.039 BAO M T, PENG J, CHEN Q G. Research progress of biodegradation of polyacrylamide by microorganisms [J]. Chemical Industry and Engineering Progress, 2011, 30(9): 2080-2086(in Chinese). doi: 10.16085/j.issn.1000-6613.2011.09.039
[3] LIN Z, LI P T, HOU D, et al. Aggregation mechanism of particles: Effect of Ca2+ and polyacrylamide on coagulation and flocculation of coal slime water containing illite [J]. Minerals, 2017, 7(2): 30. doi: 10.3390/min7020030 [4] ZOU W J, ZHAO J L, SUN C B. Adsorption of anionic polyacrylamide onto coal and kaolinite calculated from the extended DLVO theory using the van oss-chaudhury-good theory [J]. Polymers, 2018, 10(2): 113. doi: 10.3390/polym10020113 [5] CASTRO S, LASKOWSKI J S. Depressing effect of flocculants on molybdenite flotation [J]. Minerals Engineering, 2015, 74: 13-19. doi: 10.1016/j.mineng.2014.12.027 [6] HU H, LIU J F, LI C Y, et al. Anaerobic biodegradation of partially hydrolyzed polyacrylamide in long-term methanogenic enrichment cultures from production water of oil reservoirs [J]. Biodegradation, 2018, 29(3): 233-243. doi: 10.1007/s10532-018-9825-1 [7] ZHAO L M, BAO M T, YAN M, et al. Kinetics and thermodynamics of biodegradation of hydrolyzed polyacrylamide under anaerobic and aerobic conditions [J]. Bioresource Technology, 2016, 216: 95-104. doi: 10.1016/j.biortech.2016.05.054 [8] BAO M T, CHEN Q G, LI Y M, et al. Biodegradation of partially hydrolyzed polyacrylamide by bacteria isolated from production water after polymer flooding in an oil field [J]. Journal of Hazardous Materials, 2010, 184(1/2/3): 105-110. [9] WEN Q X, CHEN Z Q, ZHAO Y, et al. Biodegradation of polyacrylamide by bacteria isolated from activated sludge and oil-contaminated soil [J]. Journal of Hazardous Materials, 2010, 175(1/2/3): 955-959. [10] SONG T W, LI S S, WA D D, et al. Biodegradation of hydrolyzed polyacrylamide by the combined expanded granular sludge bed reactor-aerobic biofilm reactor biosystem and key microorganisms involved in this bioprocess [J]. Bioresource Technology, 2018, 263: 153-162. doi: 10.1016/j.biortech.2018.04.121 [11] SANG G L, PI Y R, BAO M T, et al. Biodegradation for hydrolyzed polyacrylamide in the anaerobic baffled reactor combined aeration tank [J]. Ecological Engineering, 2015, 84: 121-127. doi: 10.1016/j.ecoleng.2015.07.028 [12] NYYSSÖLÄ A, AHLGREN J. Microbial degradation of polyacrylamide and the deamination product polyacrylate [J]. International Biodeterioration & Biodegradation, 2019, 139: 24-33. [13] 林玉珍, 曾光明, 张娱, 等. 有机农药滴滴涕和毒死蜱生物降解机制的分子模拟研究 [J]. 环境科学, 2012, 33(3): 1015-1019. LIN Y Z, ZENG G M, ZHANG Y, et al. Biodegradation mechanism of DDT and chlorpyrifos using molecular simulation [J]. Environmental Science, 2012, 33(3): 1015-1019(in Chinese).
[14] LIU Z F, SHAO B B, ZENG G M, et al. Effects of rhamnolipids on the removal of 2, 4, 2, 4-tetrabrominated biphenyl ether (BDE-47) by Phanerochaete chrysosporium analyzed with a combined approach of experiments and molecular docking [J]. Chemosphere, 2018, 210: 922-930. doi: 10.1016/j.chemosphere.2018.07.114 [15] ZHANG Y, ZENG Z T, ZENG G M, et al. Enzyme-substrate binding landscapes in the process of nitrile biodegradation mediated by nitrile hydratase and amidase [J]. Applied Biochemistry and Biotechnology, 2013, 170(7): 1614-1623. doi: 10.1007/s12010-013-0276-1 [16] ZHAO L M, ZHANG C C, LU Z Y, et al. Key role of different levels of dissolved oxygen in hydrolyzed polyacrylamide bioconversion: Focusing on metabolic products, key enzymes and functional microorganisms [J]. Bioresource Technology, 2020, 306: 123089. doi: 10.1016/j.biortech.2020.123089 [17] KAY-SHOEMAKE J L, WATWOOD M E, LENTZ R D, et al. Polyacrylamide as an organic nitrogen source for soil microorganisms with potential effects on inorganic soil nitrogen in agricultural soil [J]. Soil Biology and Biochemistry, 1998, 30(8/9): 1045-1052. [18] KAY-SHOEMAKE J L, WATWOOD M E, SOJKA R E, et al. Polyacrylamide as a substrate for microbial amidase in culture and soil [J]. Soil Biology and Biochemistry, 1998, 30(13): 1647-1654. doi: 10.1016/S0038-0717(97)00251-4 [19] OHTAKI A, MURATA K, SATO Y, et al. Structure and characterization of amidase from Rhodococcus sp. N-771: Insight into the molecular mechanism of substrate recognition [J]. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, 2010, 1804(1): 184-192. doi: 10.1016/j.bbapap.2009.10.001 [20] ZHAO X D, SONG L Z, FU J, et al. Experimental and DFT investigation of surface degradation of polyvinylidene fluoride membrane in alkaline solution [J]. Surface Science, 2011, 605(11/12): 1005-1015. [21] TU M L, WANG C, CHEN C, et al. Identification of a novel ACE-inhibitory peptide from casein and evaluation of the inhibitory mechanisms [J]. Food Chemistry, 2018, 256: 98-104. doi: 10.1016/j.foodchem.2018.02.107 [22] CHEN M, ZENG G M, LAI C, et al. Molecular basis of laccase bound to lignin: Insight from comparative studies on the interaction of Trametes versicolor laccase with various lignin model compounds [J]. RSC Advances, 2015, 5(65): 52307-52313. doi: 10.1039/C5RA07916K [23] 刘文强, 黄露义, 李国菠. 采用药效团模型和分子对接方法筛选新型的组蛋白甲基转移酶G9a抑制剂 [J]. 西北药学杂志, 2016, 31(2): 186-189. doi: 10.3969/j.issn.1004-2407.2016.02.022 LIU W Q, HUANG L Y, LI G B. Virtual screening of novel histone methyltransferase G9a inhibitors by using pharmacophore modeling and molecular docking [J]. Northwest Pharmaceutical Journal, 2016, 31(2): 186-189(in Chinese). doi: 10.3969/j.issn.1004-2407.2016.02.022
[24] TU M L, LIU H X, ZHANG R Y, et al. Analysis and evaluation of the inhibitory mechanism of a novel angiotensin-I-converting enzyme inhibitory peptide derived from casein hydrolysate [J]. Journal of Agricultural and Food Chemistry, 2018, 66(16): 4139-4144. doi: 10.1021/acs.jafc.8b00732 -



 下载:
下载: