-
多氯代二苯并-对-二恶英和多氯代二苯并呋喃(PCDD/Fs)是传统的持久性有机污染物(POPs),具有POPs典型的持久性、高毒性、长距离迁移性和生物累积性的特征[1],能给动物和人类带来严重的健康风险。随着全球工业的快速发展,PCDD/Fs排放源的数量不停增长密度持续增大,农产品也不可避免的受到了影响[2]。从20世纪末到21世纪,PCDD/Fs污染畜禽饲料及农产品等的污染事件频繁发生[2-3]。华北某地区是著名的“钢都”同时也是全国第二大的“奶都”。钢铁厂是PCDD/Fs污染的典型来源,钢铁厂坐落在农田附近又紧挨奶牛养殖场,农田中农作物的生产不可避免的会受到影响。而相当部分的农作物会作为畜禽动物的饲料原料直接进行回收加工,使得PCDD/Fs沿着食物链进行传递。目前关于PCDD/Fs的研究主要集中在环境污染调查和致毒效应探究[4]。并且国际上关于奶牛这种大型哺乳动物的污染物暴露实验极少,多数实验采用纯品添加进饲料进行暴露,无法还原实际污染条件,可能存在夸大PCDD/Fs暴露风险的问题。
代谢组学是研究生物内源性小分子代谢物整体及其变化规律的学科,以高通量、高灵敏度著称[5],是继基因组学和蛋白组学新发展起来的学科。近年来代谢组学的发展为毒理学的深入研究提供了新的技术手段和思路。可以通过检测代谢通路上多种代谢物的变化来探究代谢紊乱背后的机制,评价环境污染物暴露所带来的毒性效应[6],进而推断毒性作用的分子机制,具有快速和高灵敏度等特点。血液作为机体的重要媒介体液,包含了不同组织器官的多种代谢产物[7]。Tian等[8]采用血液代谢组学检测与奶牛热应激有关的生物标志物,共鉴定出41种代谢产物。迄今为止对奶牛血液代谢物的研究主要是对血液中葡萄糖、胆固醇、游离脂肪酸等常规参数的测定,很少有研究探究污染物对奶牛整体代谢的影响。
本研究通过在饲料中添加飞灰暴露奶牛,探究其对奶牛血液代谢组的影响,筛选差异代谢物,揭示PCDD/Fs暴露的健康效应和在奶牛体内的迁移代谢规律,寻找潜在的生物标志物。
-
场地设置在一个集中式的奶牛养殖场,选择体重约为600 kg,二胎次的荷斯坦奶牛4头进行暴露,洁净饲料饲喂奶牛为对照。由于更换养殖场地可能会导致奶牛出现严重应激反应,影响奶牛食欲和产奶量。因此适应期阶段使用洁净饲料进行饲喂,让奶牛适应环境后开始实验。将飞灰作为PCDD/Fs的载体,每日将30 g飞灰与40 kg洁净饲料充分混匀,暴露量为0.61 pgTEQ·g−1,连续饲喂38 d。于第38天采集血液,离心,收集上层血清,置于−80 ℃冰箱保存,样品信息见表1。
-
样品解冻后,取适量血清(200 μL),加3倍量乙腈(600 μL),涡旋2 min,13000 r·min−1离心10 min,取上清400 μL,氮气吹干。吹干样品后用100 μL 50%乙腈复溶,涡旋60 s,离心10 min(13000 r·min−1 ,4 ℃)。吸取上清2 μL进样。另取所有样品适量,等量混合,为QC样品。检测过程中每4个样品插入一针QC,用于监测仪器与方法的稳定性。
-
采用SCIEX Exion LC 联合 X-500R Q-TOF mass spectrometer (AB Sciex, Foster City, CA, USA)液质联用仪按表2中参数进行分析,电喷雾离子源ESI,正负离子扫描模式,扫描范围m/z100-1250。
所使用色谱柱为Waters Acquity BEH C18 column (2.1 mm×100 mm, 1.7 μm),洗脱程序如表3所示。柱温为35 ℃,流动相为A:水(含0.1%甲酸);B:乙腈;流速:0.3 mL·min−1;进样体积:2 μL。
-
通过SCIEX OS Analytics提取原始图谱并进行数据矩阵的转换,主要包括质荷比(m/z)和保留时间(Rt)及峰面积(intensity)等信息。所有数据用总峰面积进行归一化后生成的excel表用于代谢组学分析。为减少偶然误差产生的信号干扰,对QC中RSD≥40%的变量先行在excel中剔除。将excel文件导入SIMCA 14.1软件中进行多元数理统计分析。
-
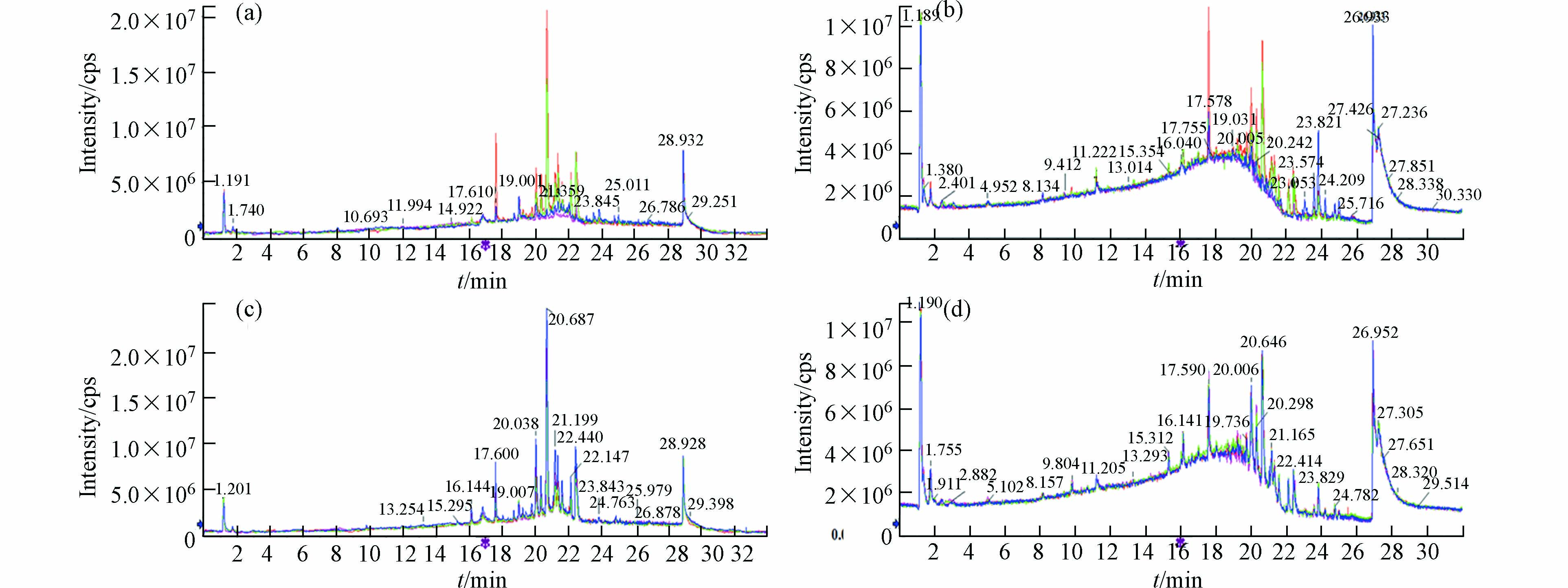
图1为对照组和暴露组正负离子模式下的叠加离子流图,可以看到保留时间和峰面积重叠较好,图谱几乎完全重合。说明在样本检测序列中,仪器方法稳定,所得数据质量可靠。
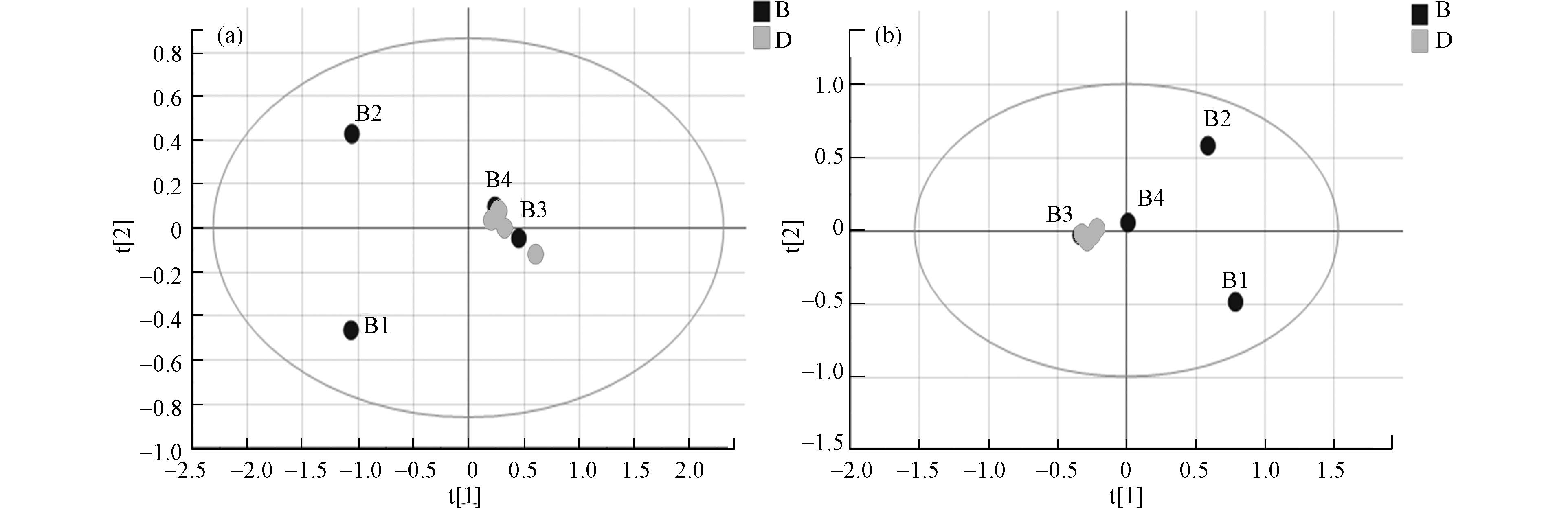
数据经预处理后进行多维统计分析和模式识别,无监督的主成分分析(PCA)结果如图2所示。该数据用于评价样品组内差异情况,点分布越聚集表示代谢组越相似。反之,分布越分散表示差异越大。结果可见, 两组样本间可以明显区分,暴露组样品分布较离散,组内差异较大。暴露组离子流图谱中也可观察到组内色谱峰组成、分布与强度有一定偏差(图1a, b)。对照组平行性较好,图谱基本一致,在PCA中也聚集紧密。以上结果提示污染物的暴露对奶牛血液代谢组产生了显著的影响。
-
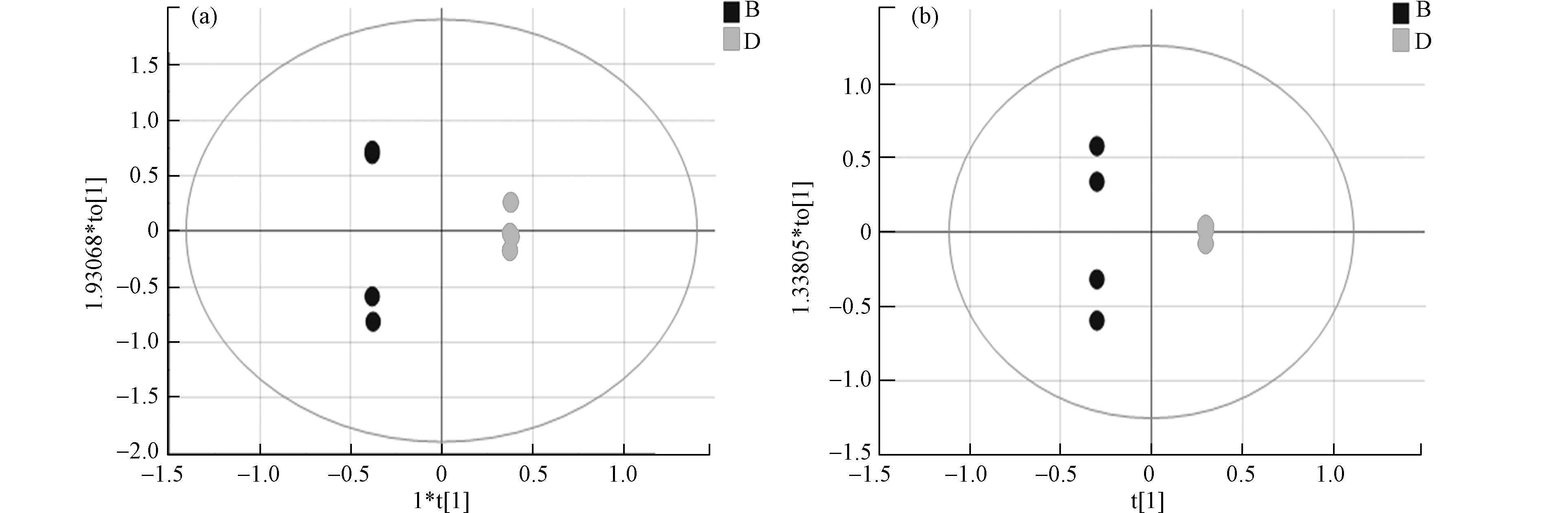
采用正交偏最小二乘法判别分析(OPLS-DA)使对照组与暴露组差异扩大,筛选差异代谢物。OPLS-DA将分组纳入变量,放大了组间差异,主要用于观测不同组间的差异大小。散点越接近说明两组间差异越小,反之则组间差异越大。图3为两组的OPLS-DA图,暴露组和对照组在正负离子模式下都有较为明显的分离趋势,正离子模式下R2X 0.984 、R2Y 1.000、Q2 0.771 ,负离子模式下R2X 0.950、R2Y 1.000、Q2 0.830, R2Y和Q2的数值均大于0.5,这表明模型预测能力强且可靠。
-
在OPLS-DA模型的基础上,根据变量投影重要性指标(VIP值)筛选差异变量(VIP>1),考虑组内差异的影响,降低pcorr标准(|pcorr|>0.4)。对筛选出的代谢物进行鉴定,因为样本量较少,故采用U检验进行补充分析,满足其中一种检验方式(P<0.05),视为差异性代谢物。最终鉴定的结果见表4,共鉴定差异代谢物50个。主要为溶血磷脂类、脂肪酸类,可从脂质代谢异常切入研究。本实验基本呈现出脂肪酸与氨基酸升高,溶血磷脂降低的结果,提示奶牛在二恶英类物质暴露下可能导致脂质代谢的紊乱,如脂肪酸的堆积等,从而进一步影响能量代谢。
-
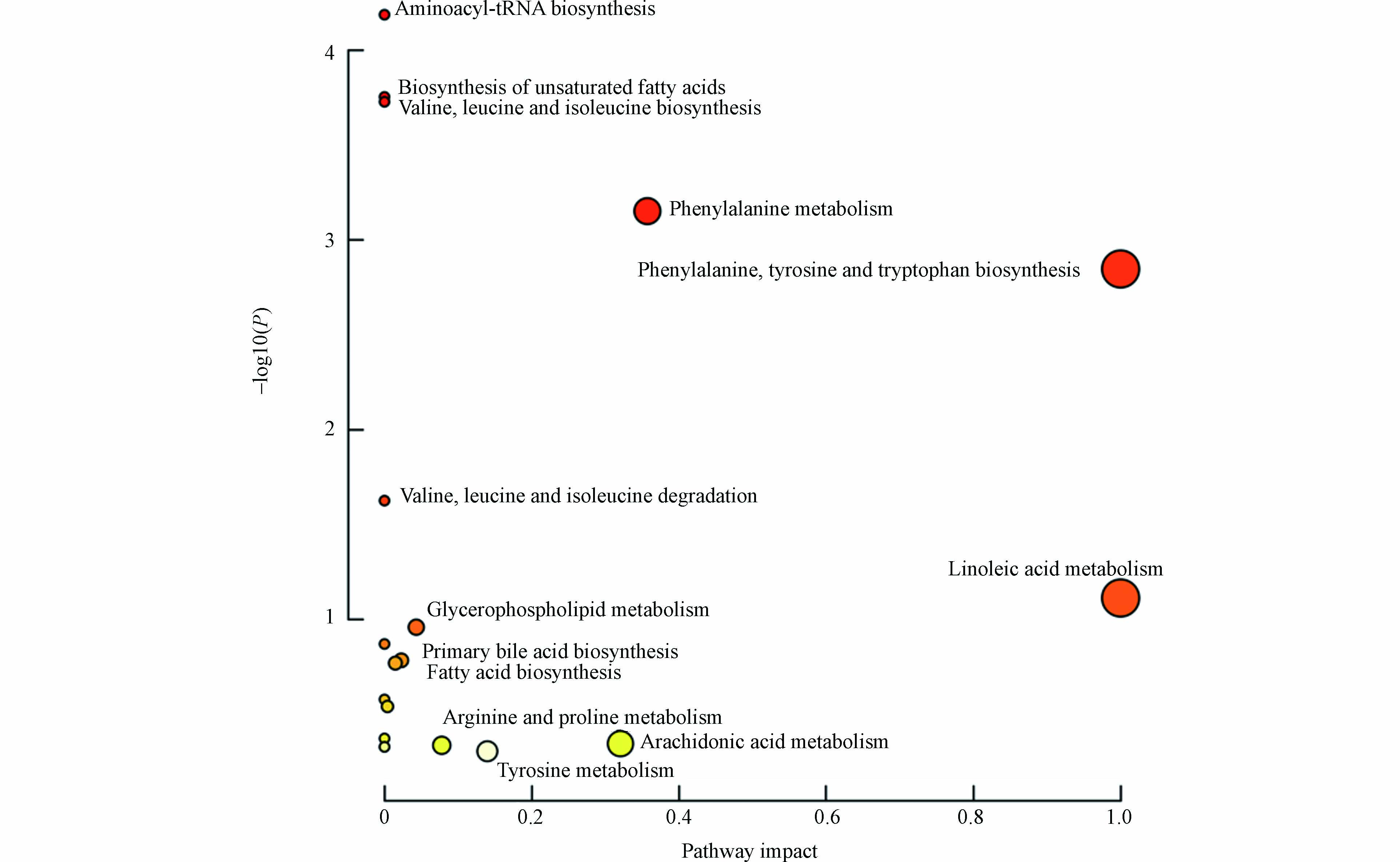
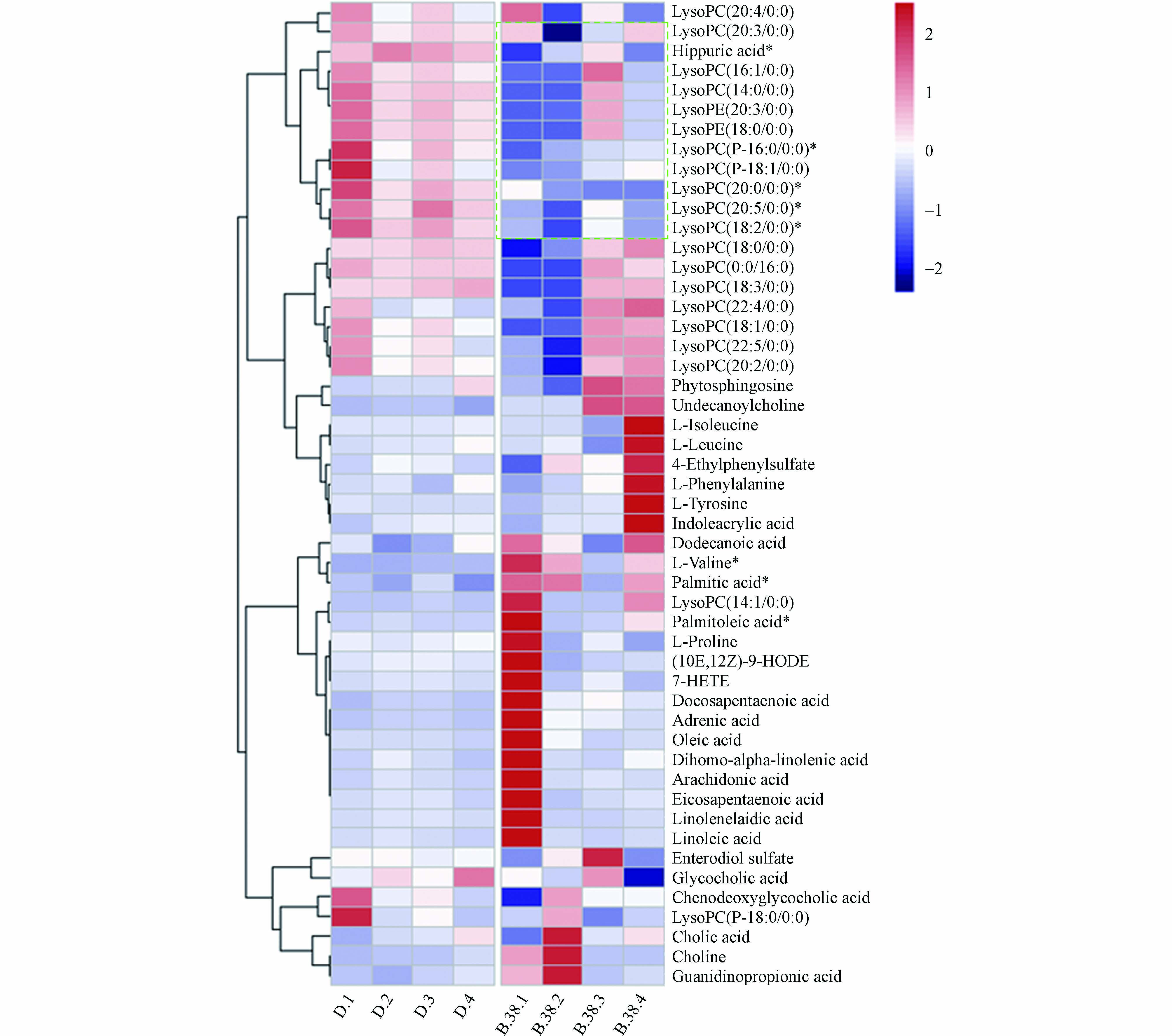
将差异化合物导入MetaboAnalyst 5.0 (http://www.MetaboAnalyst.ca/) 进行通路分析,代谢通路信息见表5,并且形成拓扑图(图4),共涉及到19条通路,主要与氨基酸、脂肪酸、胆汁酸与磷脂相关。热图见图5,颜色越红表示该代谢物响应越高,越蓝表示响应越低,可见溶血磷脂类成分在暴露后响应降低较为明显。
苯丙氨酸,酪氨酸和色氨酸均为芳香族氨基酸。芳香族氨基酸合成途径中底物均为莽草酸。其中苯丙氨酸与酪氨酸的结构相似,在体内苯丙氨酸可被苯丙氨酸羟化酶催化引入羟基生成酪氨酸[9-13]。色氨酸和苯丙氨酸均为人体无法合成的必需氨基酸,这些氨基酸血清水平的增加可能与分解代谢途径改变有关[14]。苯丙氨酸可转化生成苯丙酮酸,苯丙酮酸大量堆积会导致中枢神经系统受到损伤[15-16]。并且有研究发现动脉粥样硬化患者的心血管疾病发生风险与苯丙氨酸负相关,心绞痛患者发生短暂缺血时体内苯丙氨酸也显著降低[17]。酪氨酸是一种非必须氨基酸,可在肾上腺髓质中合成肾上腺素、去肾上腺素和多巴胺[18]。酪氨酸是黑色素的前体,临床发现炎症性皮肤病与黑色素细胞功能改变密切相关[19]。黑色素生物合成途径的第一步是酪氨酸酶催化,酪氨酸酶活性的降低会导致体内酪氨酸水平的增高[19]。苯丙氨酸、酪氨酸代谢的稳定对机体维持正常生理机能至关重要。本研究发现,与对照组相比暴露组苯丙氨酸、酪氨酸含量均显著上升,说明PCDD/Fs暴露会干扰苯丙氨酸、酪氨酸生物合成的相关代谢通路。
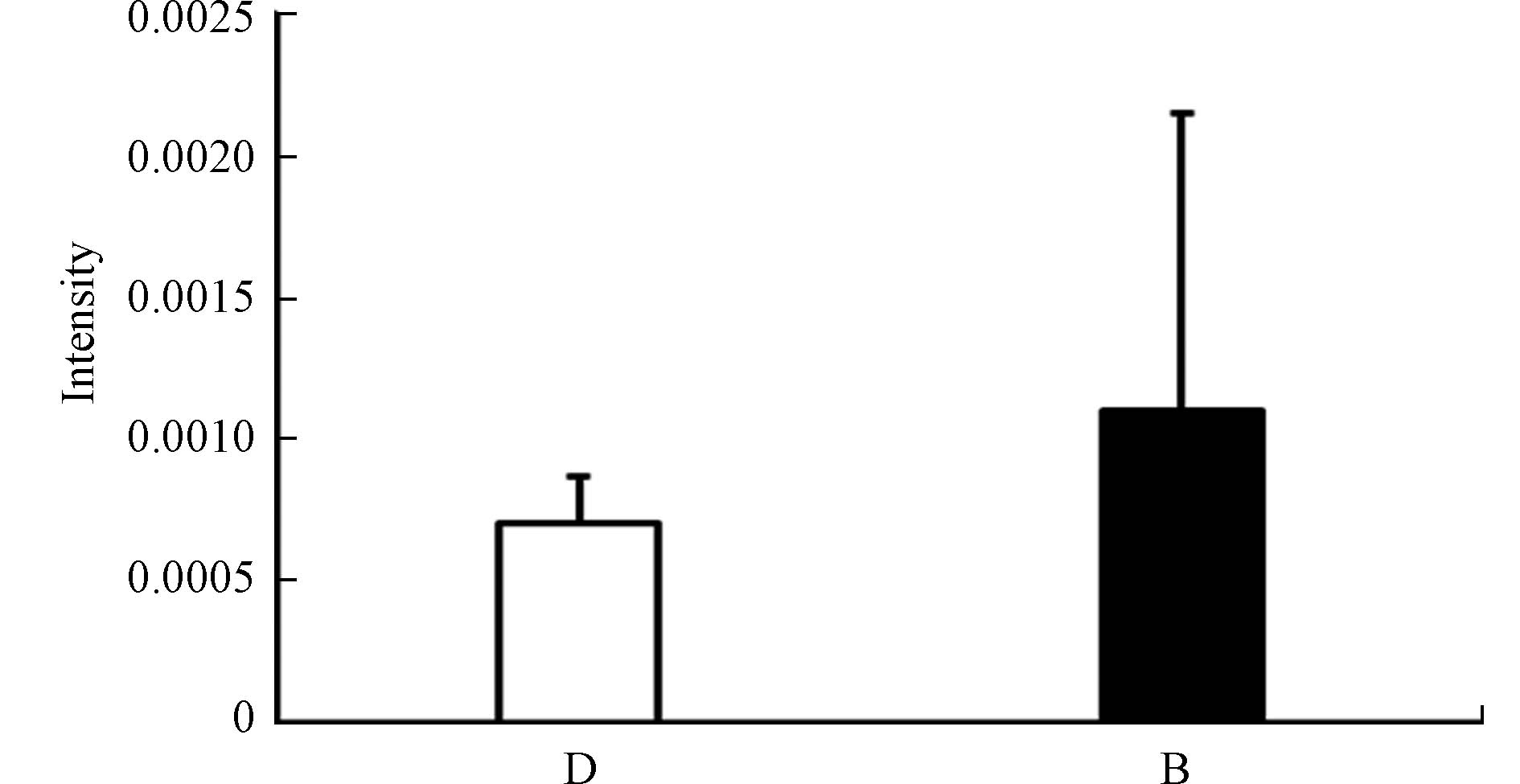
色氨酸是人体必需的10种氨基酸之一,是构成机体蛋白质的主要成分,能促进T淋巴前体细胞分化为成熟的T淋巴细胞[12]。色氨酸在体内主要有两种代谢途径:甲氧基吲哚途径和犬尿氨酸途径。犬尿氨酸途径是色氨酸非蛋白代谢的主要途径[14]。色氨酸通过犬尿氨酸途径的改变在癌症、阿尔茨海默症和抑郁症等疾病中都有过报道[14, 20-22]。有研究发现体内色氨酸水平降低会抑制T细胞的活化与增殖,使T细胞在G1期停滞,导致机体免疫功能降低[23]。甲氧基吲哚途径中包含了许多具有神经活性物质的代谢物,如5-羟色胺,褪黑素,血清素等[24-25]。有研究表明,皮肤中的血清素可以调节免疫反应,通过控制免疫细胞的凋亡导致慢性特应性皮炎的发生[25]。高苯丙氨酸水平会抑制5-羟色胺脱羧酶的活性,从而降低血清素的生成[26]。本研究中观察到色氨酸响应水平升高可能反映了色氨酸转化途径受到了干扰。在鉴定出的代谢物中,吲哚丙烯酸(Indoleacrylic acid)在暴露后浓度升高(图6),且吲哚丙烯酸为芳香烃受体(AhR)的配体。AhR信号通路与脂肪酸和氨基酸代谢高度相关,脂肪酸的积累是通过AHR-PPARA/G-FATP1信号通路介导的,可能为PCDD/Fs通过AhR影响全身代谢提供依据[9],也提示我们PCDD/Fs可能通过干扰色氨酸代谢引起免疫和神经系统的损伤。
亚油酸是生长发育过程中所必需的一种饱和脂肪酸[27]。作为前列腺素的前体,同时也是游离脂肪酸的重要成分,在基因表达调节、细胞的膜构建及细胞信号传导等方面都有至关重要的作用[27-29]。有研究表明亚油酸有助于降低血清胆固醇抑制动脉血栓的形成[30]。也有研究发现亚油酸诱导机体产生大量活性氧物质,引起机体红细胞的急性损伤[31],亚油酸可通过TLR4-NF-κB通路调控牛乳腺上皮细胞的抗乳腺炎作用,促进IL-1β、IL-6等促炎细胞因子的分泌[32]。本研究发现,暴露组的血清亚油酸响应水平显著降低,提示我们 PCDD/Fs暴露奶牛体内亚油酸代谢紊乱。
-
综上所述,本实验采用代谢组学技术研究了饲喂奶牛含PCDD/Fs的饲料血液代谢组学变化情况,发现PCDD/Fs暴露导致奶牛的脂肪酸代谢、氨基酸代谢、糖代谢均受到影响,从代谢层面揭示了PCDD/Fs的潜在健康风险。本研究共鉴定50个差异代谢物,其中包含AhR内源性配体,为探究PCDD/Fs暴露的健康风险生物标志物提供了重要依据。
二恶英暴露下奶牛血液代谢组学研究
Metabolomics study on blood of cow exposed to PCDD/Fs
-
摘要: 旨在研究食用被PCDD/Fs污染的饲料对奶牛血液代谢组的影响。选择飞灰作为PCDD/Fs的载体,暴露期间将30 g飞灰与40 kg奶牛饲料均匀混合,使用配制的污染饲料饲喂奶牛,连续饲喂38 d后采集血液。采用液相色谱-质谱(LC-MS)联用技术对血液进行代谢组学分析,并且结合主成分分析(PCA)和正交偏最小二乘法判别分析(OPLS-DA)对代谢轮廓进行模式识别分析并筛选差异代谢产物。共鉴定差异代谢物50个,主要为氨基酸、胆酸、磷脂与脂肪酸类成分。将差异代谢物进行通路分析并注释,涉及19条代谢通路,主要与氨基酸、脂肪酸及胆碱代谢相关。PCDD/Fs暴露干扰了奶牛的脂肪酸代谢、氨基酸代谢、磷脂代谢、胆汁酸代谢,可作为PCDD/Fs暴露的靶标代谢通路供进一步的研究。Abstract: The aim of this study was to explore the influence of PCDD/Fs pollution on the blood metabolism of cows. Fly ash was selected as the carrier of PCDD/Fs, 30 g of fly ash and 40 kg of cow feed were homogeneously mixed during exposure, the prepared polluted feed is used for feeding cows, and blood is collected after continuous feeding for 38 days. Using liquid chromatography-mass spectrometry (LC-MS) technology to analyze blood metabolism, principal component analysis (PCA) and orthogonal partial least squares-discriminant analysis (OPLS-DA) were combined to conduct pattern recognition analysis on metabolic profiles and screen differential metabolites. A total of 50 different metabolites were identified, mainly amino acids, bile acids, phospholipids, and fatty acids. Differential metabolites were subjected to pathway analysis and noted. There were 19 metabolic pathways involved, mainly related to the metabolism of amino acids, fatty acids and choline. In summary, the results showed that PCDD/Fs exposure interfered with fatty acid metabolism, amino acid metabolism, phospholipid metabolism and bile acid metabolism of cows, and it could be used as the target metabolic pathway of PCDD/Fs exposure for further research
-
Key words:
- PCDD/Fs /
- cow /
- metabolomics /
- LC-MS
-

-
表 1 样品信息
Table 1. Sample information
样品名称
Name数量
Quantity标记
Sample name对照组 4 D1-D4 暴露组 4 B1-B4 表 2 质谱条件
Table 2. Mass spectral condition
模式
Ion mode正离子模式
ESI (+)负离子模式
ESI (−)GS1 55 45 GS2 55 45 CUR 35 35 TEM 550 550 ISVF 5500 −4500 DP(MS/MS) 60 −60 CE(MS/MS) 35 −35 CES 15 15 表 3 流动相洗脱程序
Table 3. Mobile phase elution procedures
时间/min
Time流动相A/%
Mobile phase A流动相B/%
Mobile phase B0 95 5 8 40 60 18 3 97 21 3 97 21.1 95 5 25 95 5 表 4 差异代谢物
Table 4. Differential metabolites
差异代谢物
Identification分子式
Formula贡献度
VIP偏相关系数
p(corr)差异倍数
Fold changeP value
(T−test)P value
(U−test)Choline 胆碱 C5H14NO 3.73 0.52 2.20 0.19 0.34 Enterodiol sulfate 硫酸肠二醇 C18H22O7S 1.55 0.04 1.08 0.91 1.00 L−Valine*# L−缬氨酸 C5H11NO2 3.74 0.71 6.08 0.05 0.03 Guanidinopropionic acid 胍基丙酸 C4H9N3O2 2.27 0.54 1.66 0.16 0.20 L−Proline L−脯氨酸 C5H9NO2 1.63 0.15 1.38 0.73 0.69 L−Tyrosine L−酪氨酸 C9H11NO3 2.32 0.34 2.20 0.42 1.00 L−Isoleucine 异亮氨酸 C6H13NO2 2.56 0.25 1.69 0.55 0.34 L−Leucine L−亮氨酸 C6H13NO2 1.88 0.22 1.48 0.59 1.00 L−Phenylalanine L−苯丙氨酸 C9H11NO2 2.21 0.3 1.57 0.47 0.89 Indoleacrylic acid 吲哚丙烯酸 C11H9NO2 2.67 0.29 1.56 0.48 1.00 Glycocholic acid 甘氨胆酸 C26H43NO6 2.5 −0.41 0.70 0.31 0.34 Phytosphingosine 植物鞘氨醇 C18H39NO3 5.12 0.22 1.17 0.60 1.00 Cholic acid 胆酸 C24H40O5 1.21 0.26 1.20 0.53 0.69 Undecanoylcholine# 十一烷酰胆碱 C16H34NO2 2.27 0.67 2.52 0.07 0.03 Chenodeoxyglycocholic acid 乙酰脱氧甘氨胆酸 C26H43NO5 1.44 −0.31 0.84 0.46 0.89 LysoPC(14:0/0:0) 溶血磷脂酸(14:0) C22H46NO7P 2.1 −0.69 0.38 0.06 0.20 LysoPC(16:1/0:0) 溶血磷脂酸(16:1) C24H48NO7P 2.44 −0.49 0.52 0.22 0.34 LysoPE(20:3/0:0) 溶血磷脂酸(20:3) C25H46NO7P 1.05 −0.67 0.41 0.07 0.20 LysoPE(18:0/0:0) 溶血磷脂酸(18:0) C23H48NO7P 4.73 −0.65 0.41 0.08 0.20 LysoPC(20:5/0:0)*# 溶血磷脂酸(20:5) C28H48NO7P 3.71 −0.83 0.38 0.01 0.03 LysoPC(18:2/0:0)*# 溶血磷脂酸(18:2) C26H50NO7P 14.1 −0.83 0.41 0.01 0.03 LysoPC(20:4/0:0) 溶血磷脂酸(20:4) C28H50NO7P 2.31 −0.32 0.75 0.44 0.69 LysoPC(14:1/0:0) 溶血磷脂酸(14:1) C22H44NO7P 1.10 0.55 11.03 0.16 0.34 LysoPC(22:5/0:0) 溶血磷脂酸(22:5) C30H52NO7P 1.17 −0.22 0.85 0.61 0.89 LysoPC(0:0/16:0) 溶血磷脂酸(16:0) C24H50NO7P 26.27 −0.53 0.55 0.18 0.49 LysoPC(18:3/0:0) 溶血磷脂酸(18:3) C26H48NO7P 5.38 −0.51 0.58 0.20 0.69 LysoPC(20:3/0:0) 溶血磷脂酸(20:3) C28H52NO7P 2.99 −0.48 0.73 0.23 0.34 LysoPC(P-16:0/0:0)*# 溶血磷脂酸(16:0) C24H50NO6P 7.46 −0.74 0.66 0.03 0.03 LysoPC(18:1/0:0) 溶血磷脂酸(18:1) C26H52NO7P 9.09 −0.34 0.66 0.41 0.89 LysoPC(22:4/0:0) 溶血磷脂酸(22:4) C30H54NO7P 1.45 0.08 1.05 0.86 1.00 LysoPC(P-18:1/0:0) 溶血磷脂酸(18:1) C26H52NO6P 2.38 −0.60 0.75 0.11 0.11 LysoPC(P-18:0/0:0) 溶血磷脂酸(18:0) C26H54NO6P 1.42 −0.34 0.86 0.41 0.49 LysoPC(20:2/0:0) 溶血磷脂酸(20:2) C28H54NO7P 1.39 −0.36 0.74 0.38 0.69 LysoPC(18:0/0:0) 溶血磷脂酸(18:0) C26H54NO7P 14.11 −0.45 0.69 0.27 0.69 LysoPC(20:0/0:0)*# 溶血磷脂酸(20:0) C28H58NO7P 2.51 −0.82 0.52 0.01 0.03 4-Ethylphenylsulfate 4-乙基苯基硫酸盐 C8H10O4S 2.19 −0.28 1.20 0.50 0.34 (10E,12Z)-9-HODE 十八碳二烯酸 C18H32O3 3.40 −0.21 1.53 0.61 0.34 7-HETE 7-羟基花生四烯酸 C20H32O3 2.23 −0.31 1.81 0.45 1.00 Dodecanoic acid 十二烷酸 C12H24O2 1.02 −0.51 1.37 0.19 0.34 Eicosapentaenoic acid 二十碳五烯酸 C20H30O2 2.39 −0.35 1.88 0.39 1.00 Linolenelaidic acid 亚油酸 C18H30O2 4.10 −0.34 2.83 0.42 0.49 Palmitoleic acid 棕榈酸 C16H30O2 2.45 −0.46 3.81 0.25 0.69 Arachidonic acid 花生四烯酸 C20H32O2 7.30 −0.41 2.02 0.31 0.20 Docosapentaenoic acid# 二十二碳五烯酸 C22H34O2 2.46 −0.54 1.99 0.17 0.03 Linoleic acid 亚油酸 C18H32O2 10.71 −0.35 1.75 0.39 0.69 Dihomo-alpha-linolenic acid 亚麻脂酸 C20H34O2 4.31 −0.40 1.51 0.32 0.49 Adrenic acid# 肾上腺酸 C22H36O2 2.94 −0.53 2.83 0.18 0.03 Palmitic acid* 软脂酸 C16H32O2 4.69 −0.73 1.45 0.04 0.11 Oleic acid 油酸 C18H34O2 4.95 −0.41 1.81 0.31 0.49 Hippuric acid*# 马尿酸 C9H9NO3 5.98 0.81 0.40 0.01 0.03 *表示T检验下有显著性差异,#表示Mann-Whitney U检验下有显著性差异 表 5 代谢通路信息
Table 5. Metabolic pathway information
通路名称
Pathways相关代谢物
MetabolitesP值
-lg10(p)影响
ImpactAminoacyl-tRNA biosynthesis L-Phenyl`alanine; L-Valine; L-Isoleucine; L-Leucine; L-Tyrosine; L-Proline 4.19 0.00 Biosynthesis of unsaturated fatty acids Hexadecanoic acid; (9Z)-Octadecenoic acid; Linoleate; Arachidonate; (5Z,8Z,11Z,14Z,17Z)-Icosapentaenoic acid 3.75 0.00 Valine, leucine and isoleucine biosynthesis L-Leucine; L-Isoleucine; L-Valine 3.73 0.00 Phenylalanine metabolism L-Phenylalanine; Hippurate; L-Tyrosine; 3.15 0.36 Phenylalanine, tyrosine and tryptophan biosynthesis L-Phenylalanine; L-Tyrosine; 2.85 1.00 Valine, leucine and isoleucine degradation L-Valine; L-Isoleucine; L-Leucine 1.63 0.00 Linoleic acid metabolism Linoleate 1.11 1.00 Glycerophospholipid metabolism 1-Acyl-sn-glycero-3-phosphocholine; Choline 0.96 0.04 Ubiquinone and other terpenoid-quinone biosynthesis L-Tyrosine 0.87 0.00 Primary bile acid biosynthesis Cholic acid; Glycocholate 0.79 0.02 Fatty acid biosynthesis Hexadecanoic acid; Dodecanoic acid 0.77 0.01 Pantothenate and CoA biosynthesis L-Valine 0.58 0.00 Sphingolipid metabolism Phytosphingosine 0.54 0.00 Glycine, serine and threonine metabolism Choline 0.37 0.00 Arachidonic acid metabolism Arachidonate 0.35 0.32 Arginine and proline metabolism L-Proline 0.34 0.08 Fatty acid elongation Hexadecanoic acid 0.33 0.00 Fatty acid degradation Hexadecanoic acid 0.33 0.00 Tyrosine metabolism L-Tyrosine 0.31 0.14 Aminoacyl-tRNA biosynthesis L-Phenylalanine; L-Valine; L-Isoleucine; L-Leucine; L-Tyrosine; L-Proline 4.19 0.00 Biosynthesis of unsaturated fatty acids Hexadecanoic acid; (9Z)-Octadecenoic acid; Linoleate; Arachidonate; (5Z,8Z,11Z,14Z,17Z)-Icosapentaenoic acid 3.75 0.00 -
[1] MREMA E J, RUBINO F M, BRAMBILLA G, et al. Persistent organochlorinated pesticides and mechanisms of their toxicity [J]. Toxicology, 2013, 307: 74-88. doi: 10.1016/j.tox.2012.11.015 [2] LÜ H, CAI Q Y, JONES K C, et al. Levels of organic pollutants in vegetables and human exposure through diet: A review [J]. Critical Reviews in Environmental Science and Technology, 2014, 44(1): 1-33. doi: 10.1080/10643389.2012.710428 [3] PANEL ON CONTAMINANTS IN THE FOOD CHAIN (CONTAM) E F S A, KNUTSEN H K, ALEXANDER J, et al. Risk for animal and human health related to the presence of dioxins and dioxin-like PCBs in feed and food [J]. EFSA Journal. European Food Safety Authority, 2018, 16(11): e05333. [4] SSEBUGERE P, SILLANPÄÄ M, MATOVU H, et al. Human and environmental exposure to PCDD/Fs and dioxin-like PCBs in Africa: A review [J]. Chemosphere, 2019, 223: 483-493. doi: 10.1016/j.chemosphere.2019.02.065 [5] GRIFFIN J L. The potential of metabonomics in drug safety and toxicology [J]. Drug Discovery Today:Technologies, 2004, 1(3): 285-293. doi: 10.1016/j.ddtec.2004.10.011 [6] ZHANG J, YAN L J, TIAN M P, et al. The metabonomics of combined dietary exposure to phthalates and polychlorinated biphenyls in mice [J]. Journal of Pharmaceutical and Biomedical Analysis, 2012, 66: 287-297. doi: 10.1016/j.jpba.2012.03.045 [7] HA M H, LEE D H, SON H K, et al. Association between serum concentrations of persistent organic pollutants and prevalence of newly diagnosed hypertension: Results from the National Health and Nutrition Examination Survey 1999–2002[J]. Journal of Human Hypertension, 2009, 23(4): 274-286. [8] TIAN H, WANG W Y, ZHENG N, et al. Identification of diagnostic biomarkers and metabolic pathway shifts of heat-stressed lactating dairy cows [J]. Journal of Proteomics, 2015, 125: 17-28. doi: 10.1016/j.jprot.2015.04.014 [9] YE G Z, GAO H, ZHANG X, et al. Aryl hydrocarbon receptor mediates benzo[a]Pyrene-induced metabolic reprogramming in human lung epithelial BEAS-2B cells [J]. Science of the Total Environment, 2021, 756: 144130. doi: 10.1016/j.scitotenv.2020.144130 [10] PANFILI E, GERLI R, GROHMANN U, et al. Amino acid metabolism in rheumatoid arthritis: Friend or foe? [J]. Biomolecules, 2020, 10(9): 1280. doi: 10.3390/biom10091280 [11] EMERY P W. Amino acids: metabolism[M]//Encyclopedia of Human Nutrition. Amsterdam: Elsevier, 2013: 72-78. [12] LINGENS F. The biosynthesis of aromatic amino acids and its regulation [J]. Angewandte Chemie International Edition in English, 1968, 7(5): 350-360. doi: 10.1002/anie.196803501 [13] WALØEN K, KLEPPE R, MARTINEZ A, et al. Tyrosine and tryptophan hydroxylases as therapeutic targets in human disease [J]. Expert Opinion on Therapeutic Targets, 2017, 21(2): 167-180. doi: 10.1080/14728222.2017.1272581 [14] SCHWARCZ R. The kynurenine pathway of tryptophan degradation as a drug target [J]. Current Opinion in Pharmacology, 2004, 4(1): 12-17. doi: 10.1016/j.coph.2003.10.006 [15] DEARMOND P D, DIETZEN D J, PYLE-EILOLA A L. Amino acids disorders[M]//Biomarkers in Inborn Errors of Metabolism. Amsterdam: Elsevier, 2017: 25-64. [16] KRISHNAN A, SOLDATI-FAVRE D. Amino acid metabolism in api complexan parasites [J]. Metabolites, 2021, 11(2): 61. doi: 10.3390/metabo11020061 [17] INCALZA M A, D'ORIA R, NATALICCHIO A, et al. Oxidative stress and reactive oxygen species in endothelial dysfunction associated with cardiovascular and metabolic diseases [J]. Vascular Pharmacology, 2018, 100: 1-19. doi: 10.1016/j.vph.2017.05.005 [18] WURTMAN R J, LARIN F, MOSTAFAPOUR S, et al. Brain catechol synthesis: Control by train tyrosine concentration [J]. Science, 1974, 185(4146): 183-184. doi: 10.1126/science.185.4146.183 [19] HIRAMOTO K, KOBAYASHI H, ISHII M, et al. Increased alpha-melanocyte-stimulating hormone (alpha-MSH) levels and melanocortin receptors expression associated with pigmentation in an NC/Nga mouse model of atopic dermatitis [J]. Experimental Dermatology, 2010, 19(2): 132-136. doi: 10.1111/j.1600-0625.2009.00988.x [20] WISSMANN P, GEISLER S, LEBLHUBER F, et al. Immune activation in patients with Alzheimer's disease is associated with high serum phenylalanine concentrations [J]. Journal of the Neurological Sciences, 2013, 329(1/2): 29-33. [21] SFORZINI L, NETTIS M A, MONDELLI V, et al. Inflammation in cancer and depression: A starring role for the kynurenine pathway [J]. Psychopharmacology, 2019, 236(10): 2997-3011. [22] SCHWARCZ R, STONE T W. The kynurenine pathway and the brain: Challenges, controversies and promises [J]. Neuropharmacology, 2017, 112: 237-247. doi: 10.1016/j.neuropharm.2016.08.003 [23] MEZRICH J D, FECHNER J H, ZHANG X J, et al. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells [J]. Journal of Immunology (Baltimore, Md. :1950), 2010, 185(6): 3190-3198. doi: 10.4049/jimmunol.0903670 [24] SOGA F, KATOH N, INOUE T, et al. Serotonin activates human monocytes and prevents apoptosis [J]. Journal of Investigative Dermatology, 2007, 127(8): 1947-1955. doi: 10.1038/sj.jid.5700824 [25] MENESES A, LIY-SALMERON G. Serotonin and emotion, learning and memory [J]. Reviews in the Neurosciences, 2012, 23(5/6): 543-553. [26] HASHIM N A A, AB-RAHIM S, SUDDIN L S, et al. Global serum metabolomics profiling of colorectal cancer [J]. Molecular and Clinical Oncology, 2019, 11(1): 3-14. [27] HAGGERTY D F, GERSCHENSON L E, HARARY I, et al. The metabolism of linoleic acid in mammalian cells in culture [J]. Biochemical and Biophysical Research Communications, 1965, 21(6): 568-574. doi: 10.1016/0006-291X(65)90523-1 [28] BRIDGES R B, CONIGLIO J G. The metabolism of linoleic and arachidonic acids in rat testis [J]. Lipids, 1970, 5(7): 628-635. doi: 10.1007/BF02531342 [29] SANDERS T A, RANA S K. Comparison of the metabolism of linoleic and linolenic acids in the fetal rat [J]. Annals of Nutrition & Metabolism, 1987, 31(6): 349-353. [30] ZÖLLNER N, WOLFRAM G. Cholesterinester im plasma als parameter der linolsäureversorgung des menschen [J]. Zeitschrift Für Die Gesamte Experimentelle Medizin Einschließ lich Experimentelle Chirurgie, 1968, 146(1): 89-92. [31] PETZOLD M, MEYER U, KERSTEN S, et al. Feeding conjugated linoleic acids and various concentrate proportions to late pregnant cows and its consequence on blood metabolites of calves [J]. Livestock Science, 2014, 161: 95-100. doi: 10.1016/j.livsci.2013.12.024 [32] MA N N, CHANG G J, HUANG J, et al. Cis-9, trans-11-conjugated linoleic acid exerts an anti-inflammatory effect in bovine mammary epithelial cells after Escherichia coli stimulation through NF-κB signaling pathway [J]. Journal of Agricultural and Food Chemistry, 2019, 67(1): 193-200. doi: 10.1021/acs.jafc.8b05500 -



 下载:
下载: