-
人工合成的有机化学品(如杀虫剂、药物和各种工业化学品)在促进社会发展、改善人类生活质量方面发挥了重要作用。Wang等[1]近期统计,目前全球市场上使用的化学品数量已达35万种。这些化学品在其整个生命周期中,都可能被释放到环境中,威胁生态系统和人类健康[1-2]。具有持久性(persistence)、生物累积性(bioaccumulation)、毒性(toxicity)的化学品,已经成为影响人体与生态健康的重要风险源[3-4]。我国《新化学物质环境管理登记指南》中明确规定应当重点管控具有PBT属性的化学物质[5]。其中,生物累积是指生物从环境和膳食(含吞食低营养级生物)中积累化学物质,使其体内该化学物质的浓度超过周围环境中浓度的现象[6]。生物富集作为生物累积的类型之一,是指生物从周围环境中摄取某种化学物质,使其体内浓度超过周围环境中浓度的现象[6]。生物富集常用生物富集因子(BCF)来表征,BCF为化学物质在生物体内的浓度与其在环境介质中平衡浓度之比[7]。欧盟化学品注册、评估、许可和限制(REACH)法规规定,BCF是筛查生物累积性物质的重要指标之一[8]。
鱼类是水生态系统的关键物种,其体内污染物的积累程度对其他生物、甚至人类健康具有重要影响[9]。传统上,鱼体BCF的测定,可遵循经济合作与发展组织(OECD)发布的“流水式鱼类生物富集测试指南(OECD指南305)”[10]。通过该方法,虽可测得一些化学品的BCF数据,但存在测试周期长、费用高、动物实验伦理等问题,无法满足对大量商用化学品进行风险管理的现实需求[9]。因此,需要发展快速高效的替代方法来获取BCF数据。
定量构效关系(QSAR)模型,作为计算毒理学技术的核心内容,可以快速高通量地获取化学品环境暴露与危害性的相关信息[11]。QSAR通过函数或映射关系将分子结构描述符(描述分子结构特征的参数)和预测终点联系起来[11]。早期BCF的QSAR预测模型,主要基于分子的理化参数、碎片参数、溶剂化参数等物理意义明确的描述符而构建,多为线性模型[12-14]。近年来,各种机器学习算法被用于QSAR模型的构建[15-18]。2019年,Miller等[19]建立并比较了24种可用于预测BCF的线性模型(如最小二乘回归、偏最小二乘回归和岭回归)和非线性模型(如随机森林、支持向量机和多层感知机),发现大多数非线性模型对BCF的预测效果比线性模型好。
随着机器学习算法不断发展,集成模型出现并得到应用。集成模型通过投票法、平均法或学习法将多个单独模型的信息整合在一起,有望产生更准确、更稳健的预测结果[20-22]。Valsecchi等[20]发现,相对于单一模型,集成模型具有减少预测不确定性、拓宽模型应用域等优点;Li等[21]发现集成模型能够增加模型多样性并减少过拟合。集成模型在预测化学品毒性方面已有应用,如鱼类半数致死浓度(LC50)和无观测效应浓度(NOEC)的集成模型等[22]。然而,关于BCF的集成模型研究还不多见。
本研究搜集整理鱼体BCF数据并构建了数据库,计算了4000多种分子描述符,选择5种机器学习算法建立了预测BCF的单一模型,进而构建了集成模型。依据OECD关于QSAR模型构建和验证的导则[23],评价了模型的稳健性和预测能力,并进行了应用域表征。
全文HTML
-
从文献[24-25]和数据库(NITE[26], CEFIC LRI[27], DSL[28], ECOTOX EPA[29], OECD Toolbox[30]和ECHA[31])中,搜集有机化学品在不同种类鱼体的BCF测定值。按以下规则对原始数据进行处理[25]:(1) 去除无机物、混合物以及金属配合物等;(2) 当BCF值不以L·kg−1为单位,不以鱼体全身测量为基础计算,或不是在OECD推荐的物种[鲤鱼(Cyprinus carpio)、虹鳟鱼(Oncorhynchus mykiss)、黑头呆鱼(Pimephales promelas)、青鳉鱼(Oryzias latipes)、斑马鱼(Danio rerio)、蓝绿鳞鳃太阳鱼(Lepomis macrochirus)、孔雀鱼(Poecilia reticulata)、三刺鱼(Gasterosteus aculeatus)]上进行测试时,则排除该值;(3) 当同一化合物有多个实测数据时,取中值,中值根据ISO16269-7规范计算得到[32];(4) 确保每个化合物都有CAS号和SMILES码与之对应。经过整理,最终得到1384种有机化学品在不同种类鱼体的BCF实测值(单位为L·kg−1)。基于线性自由能关系的QSAR原理,将BCF实测值以10为底取对数转换为lgBCF,作为预测终点[11]。
-
使用Dragon 6.0软件计算分子结构描述符,得到4885种不同类型的描述符[33]。为了使各描述符尺度处于同一数量级,对其进行标准差法标准化处理[34]。然后对描述符进行初步筛选:去掉至少有一个缺失值的描述符,去掉为常数的描述符。以筛选得到的描述符为自变量,lgBCF为因变量,使用逐步回归分析构建多元线性回归模型。去除方差膨胀因子大于5,显著性水平大于0.001的模型,确保建模描述符之间不存在多重共线性且模型具有统计学意义[35]。综合考虑经自由度调整后的决定系数和自变量个数(通常应不超过样本个数的1/5,以避免过拟合),确定用于构建QSAR模型的分子结构描述符。
-
将1384个化合物以4∶1的比例随机划分为训练集(1107个化合物)和验证集(277个化合物),训练集用于构建模型,验证集用于对模型进行外部验证,详细数据见附件。
综合考虑机器学习算法对数据的适应能力以及类型的多样性,选择普通最小二乘(OLS)[36]、支持向量机(SVM)[37]、随机森林(RF)[38-39]、梯度提升决策树(GBDT)[40]和极端梯度提升(XGBoost)[41]这5种算法,先构建预测BCF的单一模型,进而构建集成模型。使用网格搜索交叉验证来调整模型参数,确定最优模型[42]。模型信息和相关参数见附件。
使用堆叠(Stack)方法构建集成模型[21-22, 43-46]。堆叠集成模型通常包含两层,第一层使用两个或两个以上模型对终点分别进行预测,这些模型称为基学习器(Base-learner),为充分学习训练数据,基学习器一般选择非线性模型[21-22, 46];第二层只有一个模型,负责将第一层模型的预测结果进行融合,称为元学习器(Meta-learner),为降低模型过拟合风险,元学习器一般选择线性模型[22, 45]。本研究将训练好的SVM模型、RF模型、GBDT模型、XGBoost模型随机组合作为基学习器,OLS模型作为元学习器构建堆叠集成模型。
使用经自由度调整后的决定系数(
$R^2_{{\rm{adj}}} $ )、均方根误差(RMSE)以及10折交叉验证系数($Q^2_{10{\text{-}}{\rm{fold}}} $ )评价模型效果[47-48]。训练集$R^2_{{\rm{adj}}} $ , RMSE表征模型拟合优度;验证集的$R^2_{{\rm{adj}}} $ , RMSE表征模型预测能力;训练集的$Q^2_{10{\text{-}}{\rm{fold}}} $ 表征模型稳健性[35]。模型的应用域表征采用Williams图,即化合物的杠杆值(hi)对标准残差(δ)作图[35, 47]。相关计算公式详述于附件,相关计算采用Python3.7.0软件实现[49]。
1.1. 数据库构建
1.2. 分子描述符计算与筛选
1.3. 模型构建与表征
-
经检索相关文献和数据库[12-19, 24-31, 35],搜集得到1384个有机化学品在不同种类鱼体的BCF实测值(单位为L·kg−1),构建了全面的鱼体BCF数据库,其详细信息见附件。
-
经初步筛选和逐步回归分析,最终选择8个分子结构描述符(D1, D2, ···, D8)用于构建模型,其相关信息列于表1中。
-
单一模型的相关统计参数汇总于表2中。如表2所示,线性模型(OLS模型)在预测生物累积性这种复杂生物过程时误差较大,非线性模型(SVM模型、RF模型、GBDT模型、XGBoost模型)的预测效果则有较大提升。其中,RF模型的
$Q^2_{10{\text{-}}{\rm{fold}}} $ 值最大,模型最稳健;SVM模型的$R^2_{{\rm{adj}}{\text{-}}{\rm{test}}} $ 值最大,预测准确性最好。集成模型的相关统计参数汇总于表3中。如表3所示,多数集成模型的稳健性和准确性都比单一模型有提升。图1比较了集成模型和稳健性、预测性最好的单一模型的
$Q^2_{10{\text{-}}{\rm{fold}}} $ ,$R^2_{{\rm{adj}}{\text{-}}{\rm{test}}} $ 值。从图1可见,虽然Stack-6模型的$Q^2_{10{\text{-}}{\rm{fold}}} $ 值在所有集成模型中最低,但仍与RF模型的稳健程度相当;多数集成模型(除Stack-4, Stack-5, Stack-6, Stack-9模型外)的$R^2_{{\rm{adj}}{\text{-}}{\rm{test}}} $ 值高于SVM模型。如表3所示,使用不同类型基学习器的模型效果优于使用同种类型基学习器的模型效果。综合考虑各项评价指标,Stack-7模型(基学习器为SVM,RF和XGBoost模型,元学习器为OLS模型)在所有11个集成模型中表现最佳。Stack-7模型具有最高
$R^2_{{\rm{adj}}{\text{-}}{\rm{test}}} $ 和最低RMSEtest。Roy等[50]建议评估QSAR模型的预测能力还应考虑以下标准:(1) 若MAE ≤ 0.10 × TR并且MAE + 3σ ≤ 0.20 × TR,则模型具有良好的预测能力;
(2) 若MAE > 0.15 × TR或者MAE + 3σ > 0.25 × TR,则模型预测能力较差;
(3) 若不满足上述两个条件,则模型预测能力中等。
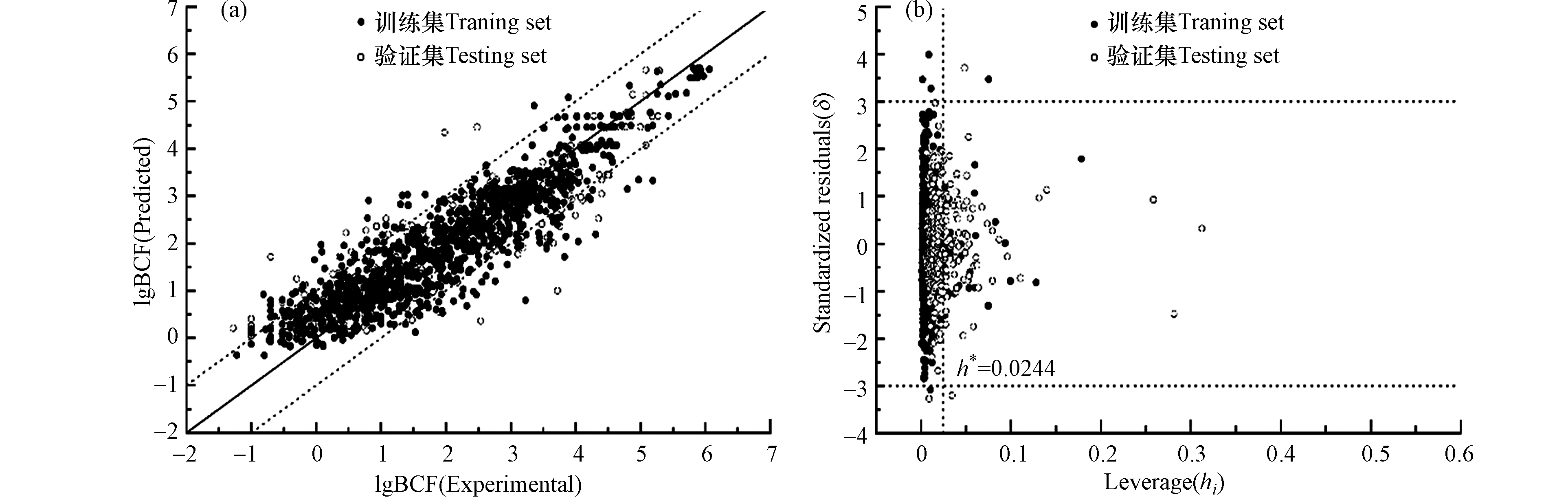
MAE表示验证集平均绝对误差;σ值表示验证集数据绝对误差值的标准偏差;TR为训练集数值范围。按此方法评价Stack-7模型的预测能力,训练集中lgBCF值范围为−1.22—6.60,TR = 7.82。剔除验证集中5%高预测误差点后,其MAE = 0.482, σ = 0.351, MAE + 3σ = 1.535,满足前述第一条标准,故Stack-7模型预测能力良好,选为最优模型,该模型的lgBCF实测值和预测值拟合图如图2a所示。
-
预测与实测值之间的差为预测残差,主要由随机误差和系统误差两部分构成[51]。随机误差由随机因素(比如训练数据的扰动)引起,具有互相抵偿性;系统误差通常来自算法本身,会造成预测结果向特定方向偏离[52]。Roy等[52]认为,如果模型满足以下条件之一,则很可能出现系统误差:
(1) nPE/ nNE > 5 或者nNE/ nPE > 5;
(2) ABS(MPE/MNE) > 2 或者ABS(MNE/MPE) > 2;
(3) AAE – ABS(AE) < 0.5 × AAE;
(4) R2(ith vs (i−1)th residuals) > 0.5;
(5) R2(Y vs residuals) > 0.5;
AE为平均残差;AAE为平均绝对残差;MPE为平均正残差;MNE为平均负残差;nPE为正残差个数;nNE为负残差个数;R2(ith vs (i−1)th residuals)表示按实测值的递增对残差进行排序,第i个残差值与第i−1个残差值之间的相关性;R2(Y vs residuals)表示预测值和残差值之间的相关性。基于上述标准对最优模型进行了评价,相关评价指标值汇总在表4中。结果表明,上述5项条件预测误差均不满足,说明最优模型不存在系统误差。
-
一般认为生物富集过程实际上是有机化合物在水相和有机相的分配过程,疏水性是生物富集过程中的主要驱动力[53],因此根据疏水性参数可以较好地预测生物富集参数。正辛醇/水分配系数(KOW)常被用于预测BCF[12-14]。Veith等[12]曾建立鱼类lgBCF与lgKOW的线性模型,模型R2 = 0.90,但模型只包含55个疏水性化合物。BLTF96是与疏水性参数KOW相关的基本描述符。本研究尝试仅使用了BLTF96描述符对数据库中1384个有机物建立线性模型,R2 = 0.40,说明仅靠疏水性这一性质难以准确估计数据库中大量化学品的lgBCF值。
表1汇总了通过逐步回归分析得到的8个分子结构描述符的含义、类型以及它们在线性模型中对应的系数。BLTF96的系数绝对值明显大于其他描述符。SpPosA_Dz(m)和ATSC7m都是与相对分子质量相关的2D描述符。Lipinski[54]发现,相对分子质量小于500的小分子药物更容易被生物体吸收。Strempel等[16]也发现相对分子质量以及分子直径对生物累积性有重要影响。综上,分子的疏水性对生物累积性影响最为显著,其次为相对分子质量和分子大小。
-
使用Williams图对Stack-7模型的应用域进行表征,以确定集成模型的适用化合物范围。如图2b所示,横坐标表示杠杆值(hi),纵坐标表示标准残差(δ)。警戒杠杆值(h*)为0.0244,认为hi ≤ h*时的化合物适用于本模型;当hi > h*时,认为该化合物超出训练集定义的描述符范围,称其为模型的应用域外化合物。模型方法的预测能力高度依赖于模型的应用域,对于应用域内的化合物预测准确性较高,而对于域外化合物的预测则存在较大不确定性[55]。当化合物的δ值落在(−3.0, +3.0)以外时,认为该点是离群点。hi > h*的化合物其δ值仍落在(−3.0, +3.0)以内,说明模型具有一定的延展性[35]。Stack-7模型的训练集和验证集中共有5个化合物(CAS号分别为81-88-9, 112-27-6, 117-80-6, 14233-37-5, 4901-51-3)的|δ| > 3且hi ≤ h*,这些化合物为模型应用域内的离群点。
2,3,4,5-四氯苯酚(CAS号:4901-51-3)、9-(2-羧基苯基)-3,6-双(二乙氨基)占吨翁氯化物(CAS号:81-88-9)的lgBCF预测值被高估。二者都含有可解离基团(酚羟基、羧基),其酸解离常数(pKa)分别为6.36和3.22[56-57]。在pH值约为7 — 9范围的水环境中,这两种物质均会以阴离子形态存在,通常离子态比其中性分子更难通过生物膜而被生物富集[58-59],所以实验测定的BCF会比仅考虑分子形态的预测值低。在将来关于BCF的QSAR预测模型构建中,应该考虑分子的解离形态[60-61]。
2,3-二氯-1,4-萘醌(CAS号:117-80-6)和1,4-二(1-异丙胺基)蒽醌(CAS号:14233-37-5)的lgBCF预测值被低估。二者作为醌类化合物,容易发生亲电加成,还原生成二元酚[62-63]。有研究发现,萘醌类化合物能够与生物亲核试剂发生反应,而生物体内的白蛋白是一种普遍存在的亲核试剂,可与含有至少一个未取代的醌碳的萘醌类化合物发生结合[64]。这类物质进入生物体后,不仅会在脂质中发生富集,还可能与蛋白质等非脂肪组织发生特定相互作用,从而造成实验测定值高于预测值的现象。
此外,有3个化合物(CAS号分别为2008-58-4, 13560-89-9, 36065-30-2)的|δ| > 3,但它们落在模型应用域外(hi > h*),因此模型对其预测的不确定性较大是可以理解的。表5列出了上述离群点以及域外化合物的分子结构、标准偏差等。
-
关于预测鱼类lgBCF的集成模型研究还较少。Zhao等[65]使用普通最小二乘回归(OLS),径向基函数神经网络(RBF-NN)和支持向量机(SVM)方法,基于473种有机化学品的lgBCF数据集建立了多个QSAR模型,并将两个使用不同描述符的RBF模型组合成一个集成模型。Gissi等[66]将两个常用预测BCF的QSAR模型按照特定规则进行了集成,其一为上述Zhao等建立的集成模型;另一为基于分子碎片的模型,该模型使用实验测得或预测的lgKOW作为唯一的描述符,并增加特定结构碎片相关的校正因子对模型进行校正[13]。
表6比较了本研究的集成模型与上述集成模型。从该表可以看出,本研究所构建的Stack-7模型在保证预测效果的基础上,应用范围更加广泛,模型表征更加严格,集成策略更加简洁,因此其在化学品BCF预测中的应用潜力更大。
-
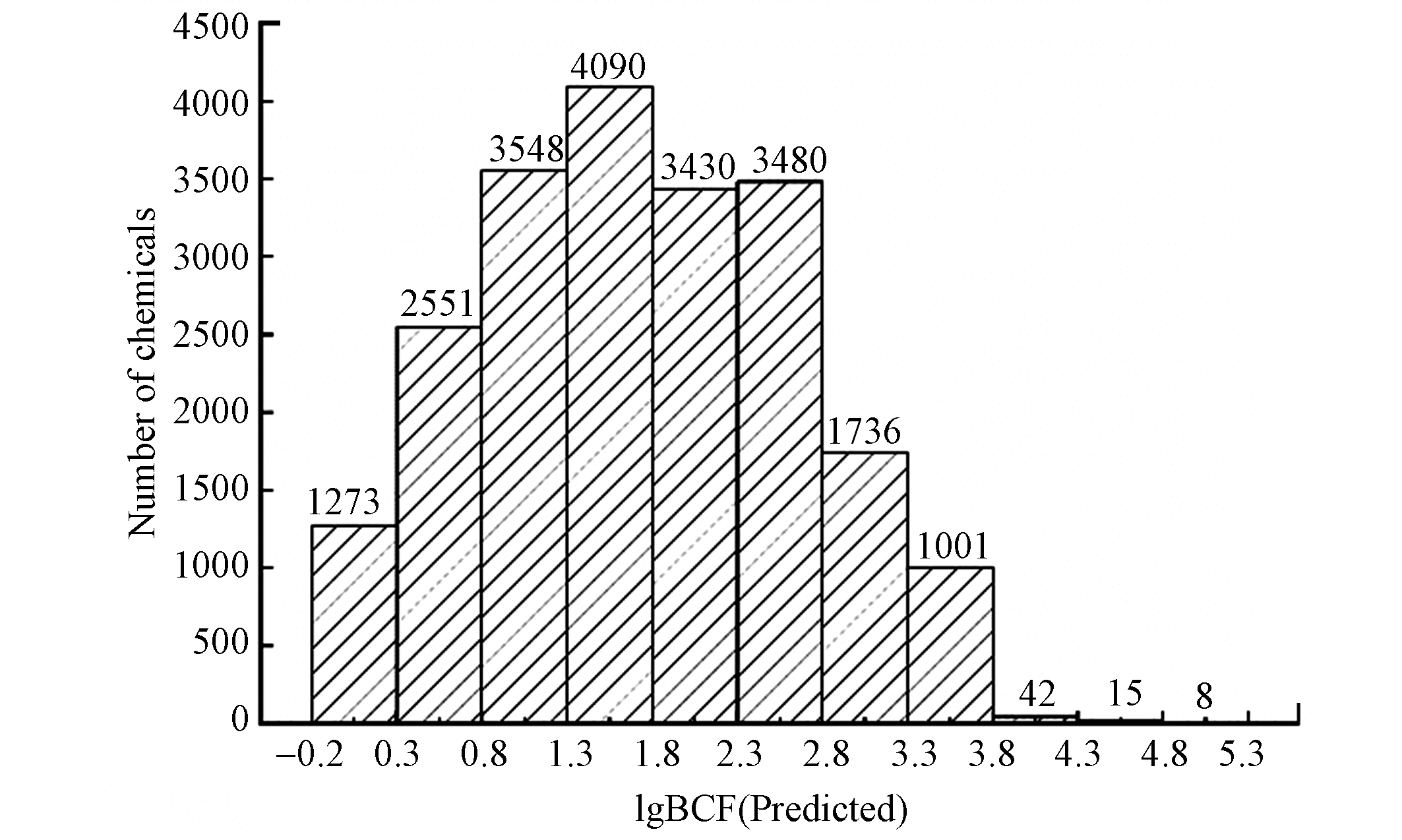
利用Stack-7模型对《中国现有化学物质名录》(IECSC)中化学物质的lgBCF进行了初步预测[67]。IECSC中21677种化学物质含有分子SMILES码,首先根据分子结构计算了所需8种描述符(BLTF96, Cl-089, SpPosA_Dz(m), SpMax1_Bh(s), B07[C-C], F02[C-O], B04[O-Cl], ATSC7m)的数值,然后计算了每种化学物质的hi值,确定有21174种在Stack-7模型应用域范围内。一般认为,当BCF ≥ 2000(即lgBCF ≥ 3.3)时,该物质具有生物累积性;BCF ≥ 5000(即lgBCF ≥ 3.7)时,具有强生物累积性[68]。IECSC中21174种化学物质的lgBCF预测值分布如图3,其中1066种化学物质具有生物累积性,86种化学物质具有强生物累积性,该预测结果可为化学品风险评价与管理工作提供参考。IECSC中化学品lgBCF预测值具体数据见附件。
2.1. 描述符筛选结果
2.2. 模型构建结果
2.3. 最优模型误差分析
2.4. 机理分析
2.5. 应用域表征
2.6. 模型比较
2.7. 模型应用
-
本研究使用OLS, RF, SVM, GBDT和XGBoost建立了预测有机化学品鱼体BCF的QSAR模型,并进一步构建了堆叠集成模型。依照QSAR模型构建和验证导则,对集成模型进行了评价和应用域表征。结果表明,集成模型比单一模型的预测准确性更高,更稳健;相较以往研究,本研究所建集成模型应用域更广泛。按照我国《新化学物质环境管理登记指南》中关于QSAR模型构建和使用的要求,进一步利用集成模型对《中国现有化学物质名录》中两万余种化学物质的lgBCF值进行了初步预测,预测结果可为化学品风险评价与管理工作提供参考。此外,本研究还建立了关于有机化学品鱼类BCF实测值数据库,有助于后续相关研究和应用工作的开展。




 下载:
下载: