-
抗生素是一类能抑制或杀灭微生物的化学物质,被广泛应用于人类和动物疾病预防和治疗[1]. 据全球药品销售数据库(IQVIA MIDASTM)统计,全球76个国家2000年至2015年间抗生素的使用量增长了65%,其中印度、中国和美国是全球最大的3个抗生素使用国,如果不加以控制,预计2030年抗生素的使用量将增长200%[2]. 抗生素被人和动物摄入后无法完全代谢,约30%—90%会以母体或活性代谢产物的形式随粪便和尿液排出体外,直接或间接进入受纳环境[3]. 研究发现,目前在用的大多数污水处理工艺对抗生素的去除能力有限,大量未经去除的抗生素随污水处理厂出水和活性污泥释放进入受纳水土环境[4]. 氟喹诺酮类、磺胺类及大环内酯类等抗生素在多种环境介质中频繁检出,其在污水处理厂出水、地表径流、地下水中的检出浓度可高达µg·L−1级,在土壤和沉积物中的检出浓度可高达mg·kg−1级[5 − 6]. 环境中残留的抗生素可对生物体产生不同程度的毒害作用,进而影响生态系统结构和功能的稳定性[7]. 此外,抗生素的环境残留还会诱导耐药性产生、增加耐药菌丰度和多样性、加速耐药基因和耐药性传播扩散,并进一步通过食物链(网)传递对人类和环境健康造成潜在威胁[8 − 9]. 近期,《柳叶刀》发表了对抗菌剂耐药性(antimicrobial resistance,AMR)全球影响的系统综述,强调AMR已成为一个全球性的健康威胁,亟需优化抗生素的使用并加强抗生素的污染控制[10]. 2021年,我国生态环境部已将抗生素列入《重点管控新污染物清单(2021版)》,“十四五”期间国家和地方生态环境部门将着手开展抗生素等新污染物的环境监测、风险评估和污染治理行动.
近年来,微藻介导的废水处理技术,因具有成本低、高效和环境友好等独特优势,日益引起广泛关注[11 − 14]. 研究证实,多种微藻具有降解抗生素的潜力,但其对部分难生物降解抗生素的去除效果有限[13, 15 − 16]. 通过添加合适碳源,构建基于微藻的共代谢体系可以促进难生物降解抗生素的有效去除[17 − 18]. 此外,还有研究发现微藻–细菌共生体系也可有效提高抗生素等污染物的去除效率[13, 15]. 而在自然水体中,微藻和细菌共生现象非常普遍[19],它们是否有助于抗生素等新污染物的自然削减也值得关注. 目前针对微藻–细菌共生体系在污水处理中的应用已展开了一些理论和应用研究[19],并在实验室水平和中试水平证实其技术上的有效性和可行性[20 − 21]. 本课题组前期也开展了斜生栅藻(Scenedesmus obliquus)、蛋白核小球藻(Chlorella pyrenoidosa)和尖细栅藻(Scenedesmus acuminatus)等多种淡水绿藻对激素、双酚化合物和抗生素等典型有机污染物的降解转化机理研究[18, 22 − 23],发现多种微藻具有降解有机污染物的潜力;此外,通过添加合适碳源构建微藻共代谢体系可以有效提高磺胺甲噁唑的去降解效果,并揭示了其降解转化规律、降解产物和路径. 同时我们也发现通过构建微藻–细菌共培养体系能有效促进抗生素降解,并结合转录水平和代谢水平等多组学技术正在开展微藻–细菌协同降解抗生素的机理研究. 基于已有研究进展,本文总结了微藻–细菌协同废水处理技术在抗生素去除中的研究结果,重点归纳了微藻–细菌共生协同降解抗生素可能的机理及其共适应机制,并对未来微藻–细菌共生体系的研究重点和发展方向进行展望.
-
近年来,基于微藻–细菌共生体系在污水处理上的诸多优点,已有大量研究围绕微藻–细菌共生体系提高废水中抗生素的去除效率展开. 表1总结了几种不同的微藻–细菌共生体系对典型抗生素的去除效果,并将其与纯藻类或纯菌类体系的研究结果进行对比. 由表1可知,与单一微藻和单一细菌处理组相比,微藻–细菌共生体系对磺胺类、大环内酯类、喹诺酮类以及四环素类等多种抗生素的去除率更高,最高可达100%,表明微藻–细菌协同更有利于对抗生素的去除. 研究表明,低浓度的抗生素不仅可以作为微藻的营养物质,还具有刺激微藻生长相关酶的分泌、促进细胞内总蛋白含量的增长等作用,因此藻类吸收利用低浓度抗生素的同时,降低环境中抗生素含量,以达到去除抗生素的目的[24]. 然而,高浓度的抗生素会对微藻产生一定毒性效应,导致微藻对抗生素的去除效率下降,而细菌的存在会减弱抗生素对微藻的毒性效应[25]. 同时,微藻也可降低抗生素对细菌的毒性效应,提高细菌的生物活性,从而提高其对抗生素的去除效果[26 − 27]. 通过微藻和细菌联合培养,不仅能提高微藻和细菌对抗生素的耐受性,还能提高抗生素的去除效果. Villar-Navarro等[28]利用高效率藻塘(high rate algae pond,HRAP)处理城市污水中的33种抗生素,发现HRAP对抗生素的平均去除率比活性污泥高5%–50% . Hom-Diaz等[29]发现厕所废水中85%的红霉素可通过微藻–细菌共生光生物反应器去除,而Ja´en-Gil等[30]利用纯培养的小球藻(Chlorella vulgaris)和莱茵衣藻(Chlamydomonas reinhardtii)对厕所废水中红霉素的去除率仅为33%. Prosenc等[31]研究发现,红霉素和克拉霉素在微藻–细菌共生系统中的半衰期分别为1.49 d和1.89 d,而在纯小球藻系统中半衰期则明显更长,分别为4.98 d和7.60 d. 另外,微藻–细菌共生体系对克拉霉素的去除率也比小球藻系统高22%(表1). 由此可见,相比于单一微藻体系,微藻–细菌共生体系更有利于废水中抗生素的去除.
-
如图1所示,微藻–细菌共生体系对抗生素的去除机理主要包括生物吸附、生物积累和生物降解等. 一般而言,抗生素会通过与微生物表面之间的物理化学作用而被快速吸附,其中部分被吸附的抗生素会缓慢通过微生物细胞壁向细胞内迁移并累积,积累在细胞内的抗生素进一步可被降解转化[41].
-
抗生素的生物吸附(biosorption)是通过吸附能力直接从废水中去除抗生素的物理化学过程[42]. Kiki等[43]研究发现,雨生红球藻(Haematococcus pluvialis)、四尾栅藻(Selenastrum capricornutum)、羊角月牙藻(Scenedesmus quadricauda)和小球藻(Chlorella vulgaris)均能有效降解抗生素,且它们对抗生素的去除具有化合物特异性. 4种藻对大环内酯类、磺胺类和氟喹诺酮类抗生素的吸附率分别在2%—7%、1%—4%和1%—3%之间. 微藻对抗生素的吸附作用大小与化合物辛醇-水分配系数(lg Kow)密切相关. 前期大量研究证实,磺胺甲噁唑(lg Kow = 0.89)、羟氨苄青霉素(lg Kow = 0.87)、左氧氟沙星(lg Kow = -0.39)、甲氧苄氨嘧啶(lg Kow = 0.91)等疏水性较弱的抗生素均表现出较低的生物吸附性[34, 44 − 46]. 此外,抗生素的生物吸附还与环境pH值有关. pH不仅可以影响抗生素的形态和电离程度,还可以影响吸附物的表面电荷,从而影响吸附物对抗生素的吸附效果[15]. 当pH为3—11时,活性污泥和栅藻(Scenedesmus almeriensis)–细菌共生系统的表面电荷以负电荷为主,磺胺类抗生素(pKa2 = 5—11)以阴离子形式存在,静电斥力作用导致活磺胺类抗生素的吸附效果不佳[33, 47 − 50]. Daneshvar等[51]研究发现,随着溶液pH从2增加至8时,四尾栅藻(Scenedesmus quadricauda)和四肩突四鞭藻(Tetraselmis suecica)对四环素的去除率从0%分别增加至49%和37%;而当溶液pH从8增加到10,微藻对四环素的去除效率下降. 这主要与S. quadricauda和T. suecica的表面电荷受环境pH影响,以及四环素含有碳正离子(pKa1 < 3.3)、亚甲基(pKa2 = 3.3—7.7)和碳二负离子(pKa3 > 7.7)有关[52]. 当溶液pH为2—3.3时,微藻和四环素表面电荷均为正电荷,两者间产生静电斥力,不利于微藻对四环素的吸附;当溶液pH为3.3—7.7时,四环素表面电荷为中性,微藻的表面电荷也逐渐变为中性,静电斥力逐渐缩减弱,有利于微藻对四环素的吸附;而当pH高于7.7时,微藻和四环素表面电荷均为负电荷,两者间产生静电斥力,导致微藻对四环素的吸附效率下降.
另一方面,在微藻的生存环境中,存在着与土壤“根际环境”类似的“藻际环境”. 藻际环境中有机物质丰富,能够吸引大量细菌的聚集,形成藻际微生物群落. 藻菌相互作用过程中,藻际有机质是连接藻类与细菌的枢纽,其中的一大类即是胞外聚合物(extracellular polymeric substances,EPS)[53]. EPS是指通过微藻和细菌的活性分泌、细胞表面物质的脱落、细胞的裂解和环境的吸附等作用而形成的高分子量天然聚合物[54],其成分主要包括多糖、蛋白质、核酸、脂质等[55]. 王龙飞等[56]研究了移动床生物膜反应器中的EPS对抗生素的去除效果,发现其对磺胺甲噻二唑和诺氟沙星的吸附率分别为14%和13%,而对四环素的吸附率高达88%. 与其它两种抗生素相比,四环素与EPS中蛋白质的结合较强,促进了EPS对四环素的生物吸附. EPS中的蛋白质因含有羧基、胺基和羟基等多种官能团和疏水区域等吸附位点,对抗生素的吸附起到了重要作用[54]. Zhang等[57]研究发现,好氧活性污泥、厌氧活性污泥和硫酸盐还原菌活性污泥的EPS对环丙沙星的吸附率分别为36%、24%和25%,并认为EPS蛋白质中的胺基和羧基对环丙沙星的生物吸附起到了重要作用. Song等[58]基于傅里叶变换红外光谱仪、X-光电子能谱仪和核磁共振波谱仪分析活性污泥EPS和四环素之间的相互作用,也发现羟基、羧基和胺基等官能团对四环素的生物吸附起主要作用.
-
抗生素的生物积累(bioaccumulation)是指微生物依赖不同的物理、化学和生物机制,摄取抗生素进入细胞内的代谢过程[15]. 现有研究表明,微藻–细菌共生体系中,除生物吸附外,微藻的生物累积在抗生素的去除中也发挥重要作用. Sun等[59]研究发现,磺胺二甲嘧啶可在蛋白核小球藻(Chlorella pyrenoidosa)细胞内累积,累积量可达到0.596 ng·mg−1. Xiong等[46]研究也发现,C. pyrenoidosa对左氧氟沙星的去除也是以生物累积为主. 在添加10 g·L−1 NaCl的情况下,左氧氟沙星在C. pyrenoidosa胞内的累积量可达101 μg·g−1. 然而,当抗生素在藻细胞内蓄积过量时,会诱导活性氧(reactive oxygen species,ROS)产生,诸如超氧游离基(·O2-)、过氧化氢(H2O2)、羟基(·OH)等[60 − 61],这些自由基的产生对微藻去除抗生素有利有弊. 一方面,活性氧自由基因其具有强氧化性有利于微藻对抗生素的去除. Bai等[62]研究发现磺胺甲噁唑的降解以微球藻(Nannochloris sp.)介导的光降解为主,微藻能够提供产生羟基、单线氧等ROS的光敏剂间接诱导抗生素发生光解. Ge等[63]研究发现,基于微藻介导的光降解恩诺沙星和环丙沙星的去除率分别高达82%和72%. 另一方面,活性氧自由基也会对微藻本身造成损失. ROS的强氧化特性会对微藻的脂类、蛋白质、DNA等生物大分子造成一定损伤,引起细胞损伤、诱变、甚至死亡,进而影响微藻对抗生素的去除效果[64].
-
抗生素的生物降解(biodegradation)是指微生物将抗生素作为碳源将其降解转化为小分子物质的过程[65]. 根据反应场所,生物降解可以分为胞内生物降解和胞外生物降解[14]. 胞内生物降解主要与微藻和细菌细胞内复杂的酶催化系统有关,而胞外生物降解则与EPS和胞外酶等密切相关. 微生物介导的抗生素胞内生物降解,通常涉及两类反应:I相反应和II相反应. I相反应主要指抗生素在细胞色素P450酶(Cyt P450)、细胞色素b5(Cyt b5)、NADPH-细胞色素P450还原酶(P450R)等I相酶的作用下降解转化为毒性较小的衍生物,通常涉及水解、氧化或还原反应等;II相反应主要指抗生素及其I相代谢产物在谷胱甘肽-S-转移酶或葡萄糖基转移酶的催化下,与谷胱甘肽、糖活有机酸等亲水官能团发生加合反应,以保护细胞免受氧化损伤[14]. 在胞外生物降解中,EPS不仅可以作为表面活性剂和乳化剂提高抗生素的生物利用度,还可以形成水合的生物膜基质,通过保持胞外酶靠近细胞,促使胞外酶能够代谢溶解胶体或固体形式的有机物,从而起到外部消化系统的作用[55].
多项研究证实,生物降解是微藻介导的抗生素去除技术的主要作用机制[66]. Xie等[67]研究发现,衣藻Tai-03(Chlamydomonas sp. Tai-03)对环丙沙星的生物降解率可达65%,并推测细胞色素P450酶系和谷胱甘肽-S-转移酶可能分别参与了环丙沙星的脱氟和环氧环开环反应. Peng等[68]研究发现,色绿藻(Chromochloris zofingiensis)对左氧氟沙星的生物降解率高达93%,且细胞色素P450酶系基因的表达量上调,在I相酶的作用下左氧氟沙星的哌嗪环上发生羟基化和脱羧基反应. 此外,研究证实微藻–细菌共生系统能有效提高抗生素的生物降解率. da Silva Rodrigues等[34, 40]研究发现,在微藻–细菌共生系统中磺胺甲噁唑和头孢氨苄的去除以生物降解为主导,其生物降解率分别为46%和99%. García-Galán等[36]研究发现,磺胺甲噁唑在HARP中的生物降解率约为75%,而其在固定式微藻–细菌共生体系中的生物降解率可达83%[35]. 然而,上述研究并没有明确微藻和细菌在生物降解抗生素中的具体作用机制.
-
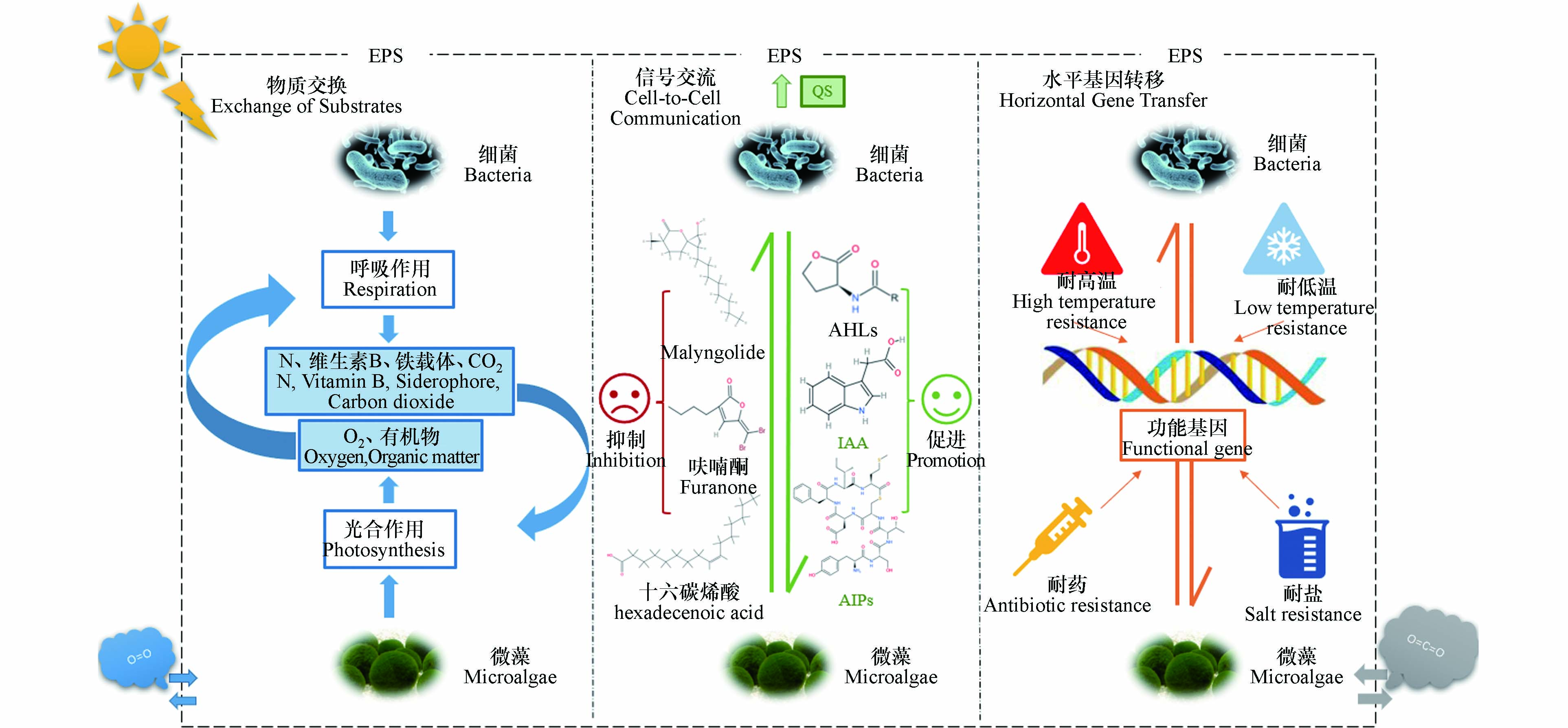
藻际环境中藻–菌互作关系复杂,包括互利、共生、拮抗和互害等多种种间关系[69]. 如图2所示,藻–菌之间的相互作用主要包括物质交换、信号交流和水平基因转移,这些相互作用在促进共生系统的形成和维持中发挥了重要的作用.
-
营养交换是微藻与细菌之间最基础的物质交换. 微藻通过光合作用等生理活动产生的O2和有机物分别可以被细菌作为呼吸作用的电子受体和底物,维持细菌的生理活动. 而细菌呼吸作用产生的固定氮、维生素B、铁载体、CO2等物质又可以维持微藻的生理活动[55, 70]. 翟春梅等[71]发现铜绿微囊藻(Microcystis aeruginosa)的细胞内含物和分泌物能促进甲基营养芽孢杆菌(Bacillus methylotrophicus,Ma-B1)的增殖. Villa等[72]将棕色固氮菌(Azotobacter vinelandii)与富油新绿藻(Neochloris oleoabundans)进行共培养,发现该细菌能为绿藻提供无机氮. Guerra-Renteria等[73]研究发现绿脓杆菌(Pseudomonas aeruginosa)能将含磷物质分解为正磷酸盐,如水解ATP的5′-核苷酸,然后将其还原成无机磷,为小球藻的生长提供磷源. 此外,微藻释放的多种有机物,如支持异养代谢的多糖和氨基酸等,也可为抗生素等有机污染物的共代谢提供充足的底物[74].
在物质交换过程中,EPS发挥了重要作用. 一方面,基于高分子聚合物组成的EPS,其相邻分子之间的孔隙充满水,聚合物包埋在水溶剂中以形成三维水凝胶网络,有利于细胞与周围介质进行物质交换[75]. 另一方面,EPS中含有多种酶,能够催化吸附于其表面的抗生素及其自身组成成分中的多糖、蛋白质等有机物分解成小分子物质,供给细菌和微藻生长利用[55, 76];此外,还能加快营养物质在微藻–细菌共生体系的传递速率. Ramanan等[77]研究发现莱茵衣藻(Chlamydomonas reinhardtii)的EPS中含有维生素依赖性蛋氨酸合成酶,该酶能协助异养细菌将维生素B12传递给微藻以维持其正常生长.
-
细菌和微藻之间的信号交流主要以群体感应(quorum sensing,QS)形式存在,QS介导的相互作用会影响微藻–细菌共生体的种间关系,改变群落结构和组成、生态系统功能,进而影响抗生素去除效果[78]. 细菌通过分泌吲哚乙酸(indole acetic acid,IAA)、自诱导肽(autoinducing oligopeptides,AIPs)、酰基高丝氨酸内酯(N-acylhomoserine laetone,AHLs)等多种QS信号分子,促进微藻胞内营养物质的加工和转化、絮凝以及油脂合成等[79 − 80]. Chen等[81]研究发现小球藻(Chlorella vulgaris)–活性污泥系统中的IAA不仅能够促进C. vulgaris生物量的增长和脂质合成,还能强化微藻与细菌之间的共生关系. Li等[82]研究发现地衣芽孢杆菌(Bacillus licheniformis)产生的AIPs通过与藻细胞膜上的感应激酶结合,激活藻细胞中叶绿素代谢基因的表达,进而促进C. vulgaris生物量的增长. 周真真[83]研究了AHLs对C. vulgaris的影响,发现AHLs不仅能调控C. vulgaris中单不饱和脂肪酸的合成,促进脂质的积累,还能调节藻细胞的抗氧化酶活性、光合代谢活性等. Wang等[84]研究发现炭疽芽孢杆菌(Bacillus anthracis)产生的AHLs能促进冈比亚藻(Gambierdiscus spp.)的生长和雪卡毒素的分泌. 此外,AHLs与EPS密切相关,AHLs的含量会影响EPS的组成成分、颗粒直径、致密程度等[85]. 微藻释放的透明胞外聚合颗粒物(transparent exopolymer particles,TEPs)、环二鸟苷酸等QS信号分子,可以调控细菌生物量增长、多糖合成等生理活动. 研究发现,赫氏圆石藻(Emiliania huxleyi)产生的TEPs能促进其共生细菌的生物量增长并强化微藻与细菌间的共生关系[86],C. vulgaris产生的环二鸟苷酸可以调控地衣芽孢杆菌(Bacillus licheniformis)多糖和AIPs的合成[87].
细菌和藻类释放的信号分子种类繁多,部分信号分子会促进对方的生理活动,而有些信号分子则会抑制对方的生理活动. 假交替单胞菌(Pseudoalteromonas sp.)的QS信号分子可激活其溶藻基因的表达,导致微藻细胞裂解[88]. 澳大利亚红藻(Delisea pulchra)能产生与AHLs 结构相似的呋喃酮,在细菌表面与受体蛋白竞争性结合,导致受体分子降解,从而抑制细菌的生长和生物膜的形成[89]. 鞘丝藻(Lyngbya spp.)产生的巨大鞘丝藻内酯可通过抑制或阻断铜绿假单胞菌(Pseudomonas aeruginosa)的LasR基因的表达,从而抑制细菌的群体感应[90].
此外,QS还与细菌的多种抗生素耐药性机制有关. Sawada等[91]研究发现,通过添加外源C4-HSL信号分子,可以上调铜绿假单胞菌的药物外排泵系统基因mexAB-oprM的表达,从而加其对多种抗生素的耐药性. 另一项研究也发现,外源添加亚精胺和精胺等QS信号分子,可以提高铜绿假单胞菌对多粘菌素B等阳离子肽类抗生素的耐药性,但同时也使其对β-内酰胺类、氯霉素、萘啶酸和甲氧苄啶等抗生素更加敏感[92]. 还有研究发现,QS信号分子能通过调控生物膜形成,形成分子或电荷屏障,以此阻止或延缓某些抗生素对细菌细胞的渗透[93 − 94].
-
水平基因转移(horizontal gene transfer, HGT)是指在差异生物个体之间,或单个细胞内部细胞器之间所进行的遗传物质的交流. 差异生物个体可以是同种但含有不同的遗传信息的生物个体,也可以是远缘的,甚至没有亲缘关系的生物个体[95]. 普遍认为水平基因转移在原核生物(细菌、古菌)间、单细胞真核生物(如微藻)间发生更为频繁,而微藻和细菌之间也会发生水平基因转移以适应极端环境[96]. 2013年发表于《Science》的研究证明细菌的一些功能基因可以转移至红藻(Galdieria sulphuraria)以助其适应高温、高盐、高酸性和高重金属残留等极端环境[97]. 此外,还有研究发现金藻(Kremastochrysopsis austriaca)耐受低温的冰结合蛋白基因是通过水平基因转移从共生细菌中获得[98]. 大多数基因水平转移发生自细菌至微藻,而细菌从微藻获取基因的概率较小. 主要有以下3点原因:1)细菌群体数量巨大,基因水平转移机会较多;2)细菌的基因及基因结构有利于其向真核生物基因组中转移;3)水平基因转移通常涉及新功能的获取,细菌在代谢上比微藻更多样化,因此它们从微藻中获得一组新的功能相关基因的可能性较小[99 − 100].
抗生素胁迫下微生物通过基因突变或基因水平转移产生和获得耐药基因、表达耐药性已获得广泛共识[101]. 研究发现,微藻对细菌耐药基因的传播扩散也有一定影响[102 − 104]. 浙工大钱海丰和中科院城环所苏建强等人研究发现,浮丝藻水华期致病菌相对丰度更高,抗生素耐药基因总量和相对丰度也显著增加,证实蓝藻水华是淡水中耐药基因扩散和富集的关键驱动因素之一[102]. 此外,还有研究发现微藻分泌物可以抑制细菌生长繁殖,阻断抗生素耐药性的传播[103 − 104]. 基因水平转移作为细菌产生耐药性的重要途径,微藻–细菌共生体系中微藻是否能通过基因水平转移获取耐药基因仍处在假设阶段,需要进一步深入研究.
-
微藻–细菌共生体系因具有生态友好、可持续发展和环境毒性耐受能力强等特性,并且能高效去除重金属、抗生素和内分泌干扰物等污染物,因此,在抗生素废水处理方面极具应用前景. 近年来,针对微藻–细菌协同废水处理技术在污水中抗生素的去除方面开展了一些基础理论和应用研究,并取得了一定的进展. 目前普遍认为,微藻–细菌协同污水处理技术在有机污染物去除和固碳方面均具有独特优势,对抗生素污染管控和碳减排都具有积极作用,已然成为未来的重要发展方向. 然而,微藻–细菌共生是一个复杂体系,现阶段对于揭示两者之间的相互作用机理仍存在许多挑战,有待进一步研究.
(1)强化微藻–细菌的协同作用机理与适用性机制研究. 目前对于微藻–细菌共生体系对抗生素降解转化的相互作用机理仍不清楚,在抗生素长期暴露下藻–菌系统的耐药性变化规律,甚至耐药基因的水平转移潜在的共适应机制仍缺乏系统研究. 因此,有必要结合高分辨质谱技术、基因组学、转录组学和代谢组学等多组学技术,从多水平分析微藻–细菌共生体系在降解转化抗生素过程中的相互作用机理和适应机制,相关研究成果有望为微藻–细菌协同技术应用和抗生素污染管控提供科学依据.
(2)筛选高效降解抗生素的微藻–细菌共生体系. 前期的研究已发现,微藻–细菌共生体系对抗生素的去除效果受多种因素影响,诸如抗生素的理化性质、环境因子、菌种、藻种等. 因此,未来有必要深入探究不同因素对抗生素去除的影响机制,通过菌种/藻种筛选、驯化等手段强化微藻–细菌共生体系对抗生素的去除能力,扩大其在抗生素污染去除中的应用.
(3)优化微藻–细菌协同废水处理技术的实际可应用性. 目前,微藻–细菌共生体系去除抗生素的研究普遍集中于实验室研究,由于实验室水平的暴露环境条件与实际废水环境存在较大差异,不足以准确评估微藻–细菌共生体系对于实际废水中抗生素的去除能力. 因此,未来应更多的开展中试和大规模水平实验,验证微藻–细菌共生体系在实际抗生素废水中的去除效果和适应性.
微藻–细菌协同去除抗生素机理及其共适应机制
Synergistic removal of antibiotics by microalgae–bacteria consortium and its co-adaptation mechanisms
-
摘要: 环境中抗生素残留不仅会影响生态系统结构和功能,还会诱导细菌耐药性的产生和传播,威胁人类健康. 我国生态环境部已将抗生素列入《重点管控新污染物清单(2021版)》,抗生素的环境监测和去除技术研发是对其污染管控的关键. 微藻–细菌共生体系具有生态友好、可持续发展和环境毒性耐受能力强等特性,可有效去除重金属、抗生素和内分泌干扰物等污染物. 鉴于微藻–细菌协同污水处理技术在提高污染物去除效率和固碳方面均具有独特优势,本文总结了微藻–细菌协同技术在抗生素废水处理中的研究和应用进展,重点归纳了微藻–细菌协同去除抗生素的可能机理及其适应机制,并对微藻–细菌共生体系未来研究重点和发展方向进行展望,以期为微藻–细菌协同污水处理技术的推广应用和抗生素污染管控提供科学依据.Abstract: The residual of antibiotics in the environment could affect the structure and function of the ecosystem, and pose potential risks to human health by inducing the formation and spread of antibiotic resistance genes. Antibiotics have been included in the list of emerging contaminants under national control by China government, and exploring effective methods for the monitor and remove of antibiotics is essential to manage and control of antibiotics pollution. Previous studies have demonstrated that microalgae-bacteria consortium could effectively remove various contaminants, such as heavy metals, antibiotics and endocrine disruptors, due to its eco-friendly, sustainable development and tolerance to environmental toxicity. Microalgae-bacteria consortium is crucial for removal of contaminants in wastewater treatment, and exhibits a great potential in carbon sequestration. Herein, we reviewed the research advances on the application of microalgae-bacteria synergistic technology in antibiotics wastewater treatment, and focused on the possible inner mechanisms of microalgae-bacteria interactions in removal antibiotics, and their co-adaptation mechanisms under antibiotics selection pressure. Then, we presented some research challenges and proposed future directions of microalgae-bacteria symbiosis system. This systematic review provides scientific basis for the promotion and application of microalgae-bacteria consortium technology, and management and control of antibiotics pollution.
-

-
表 1 不同系统对多种抗生素的去除率
Table 1. Removal rates of various antibiotics by different systems
抗生素
Antibiotics初始浓度
Initial concentration系统
Systems处理时间/d
Treatment time去除率
Removal rate参考文献
References青霉素G
土霉素
多西环素
麻保沙星
磺胺二甲基嘧啶
硫粘菌素1920 ng·L−1
22400 ng·L−1
8570 ng·L−1
713 ng·L−1
4090 ng·L−1
35 ng·L−1微藻-细菌共生光
生物反应器11 89%
93%
95%
71%
44%
89%[32] 紫色光合细菌光
生物反应器77%
—
45%
-14%
-23%
74%克拉霉素
磺胺甲噁唑2 μg·L−1
20 μg·L−1微藻-细菌共生系统 11 100%
62%[31] 小球藻(Chlorella vulgaris) 12 78%
66%四环素
环丙沙星
磺胺嘧啶
磺胺甲噁唑20、100、500、
1000 μg·L−1栅藻(Scenedesmus almeriensis)+污水细菌 4 80%、80%、62%、75%
100%、64%、46%、43%
30%、26%、36%、12%
30%、17%、21%、0%[33] 磺胺甲噁唑 52 μg·L−1 微藻-细菌共生系统 7 54% [34] 磺胺甲噁唑 500 μg·L−1 悬浮式微藻-细菌共生系统
固定式微藻-细菌共生系统7 80%
95%[35] 磺胺甲噁唑 383 ng·L−1 高效率藻塘 4.5 85% [36] 四环素 100 μg·L−1 高效率藻塘 7 99% [37] 四环素 2 mg·L−1 高效率藻塘 46 69% [38] 环丙沙星 2 mg·L−1 高效率藻塘 7 97% [39] 土霉素
恩诺沙星0.1、1、5、10 mg·L−1
0.02、0.1、1、5 mg·L−1小球藻(Chlorella vulgaris)+地衣芽孢杆菌(Bacillus licheniformis) 10 98%–100%
11%–47%[24] 头孢氨苄
红霉素50 μg·L−1 引藻(Chlorella sorokiniana)+布列文尼迪单胞菌(Brevundimonas basaltis) 7 96%
92%[40] 红霉素 (661 ± 42)ng·L−1 微藻-细菌共生光生物反应器 7 85% [29] -
[1] THAKARE R, KESHARWANI P, DASGUPTA A, et al. Chapter 1 - Antibiotics: past, present, and future [M]//P KESHARWANI, S CHOPRA, A DASGUPTA. Drug Discovery Targeting Drug-Resistant Bacteria. Academic Press. 2020: 1-8. [2] KLEIN E Y, van BOECKEL T P, MARTINEZ E M, et al. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015 [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(15): E3463-E3470. [3] WATKINSON A J, MURBY E J, COSTANZO S D. Removal of antibiotics in conventional and advanced wastewater treatment: Implications for environmental discharge and wastewater recycling [J]. Water Research, 2007, 41(18): 4164-4176. doi: 10.1016/j.watres.2007.04.005 [4] CETECIOGLU Z, ATASOY M. Biodegradation and inhibitory effects of antibiotics on biological wastewater treatment systems[M]//BIDOIA E D, MONTAGNOLLI R N. Toxicity and Biodegradation Testing. New York, NY; Springer New York. 2018: 29-55. [5] RATHI B S, KUMAR P S, SHOW P L. A review on effective removal of emerging contaminants from aquatic systems: Current trends and scope for further research [J]. Journal of Hazardous Materials, 2021, 409: 124413. doi: 10.1016/j.jhazmat.2020.124413 [6] OBEROI A S, JIA Y Y, ZHANG H Q, et al. Insights into the fate and removal of antibiotics in engineered biological treatment systems: A critical review [J]. Environmental Science & Technology, 2019, 53(13): 7234-7264. [7] YANG Q L, GAO Y, KE J, et al. Antibiotics: An overview on the environmental occurrence, toxicity, degradation, and removal methods [J]. Bioengineered, 2021, 12(1): 7376-7416. doi: 10.1080/21655979.2021.1974657 [8] LI S N, ZHANG C F, LI F X, et al. Technologies towards antibiotic resistance genes (ARGs) removal from aquatic environment: A critical review [J]. Journal of Hazardous Materials, 2021, 411: 125148. doi: 10.1016/j.jhazmat.2021.125148 [9] LIU X H, ZHANG G D, LIU Y, et al. Occurrence and fate of antibiotics and antibiotic resistance genes in typical urban water of Beijing, China [J]. Environmental Pollution, 2019, 246: 163-173. doi: 10.1016/j.envpol.2018.12.005 [10] DEREJE N. Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: A systematic analysis for the Global Burden of Disease Study 2019 [J]. The Lancet, 2020, 396(10258): 1204-1222. doi: 10.1016/S0140-6736(20)30925-9 [11] LENG L J, WEI L, XIONG Q, et al. Use of microalgae based technology for the removal of antibiotics from wastewater: A review [J]. Chemosphere, 2020, 238: 124680. doi: 10.1016/j.chemosphere.2019.124680 [12] NGUYEN H T H, MIN B. Using multiple carbon brush cathode in a novel tubular photosynthetic microbial fuel cell for enhancing bioenergy generation and advanced wastewater treatment [J]. Bioresource Technology, 2020, 316: 123928. doi: 10.1016/j.biortech.2020.123928 [13] SUTHERLAND D L, RALPH P J. Microalgal bioremediation of emerging contaminants - Opportunities and challenges [J]. Water Research, 2019, 164: 114921. doi: 10.1016/j.watres.2019.114921 [14] XIONG J Q, KURADE M B, JEON B H. Can microalgae remove pharmaceutical contaminants from water? [J]. Trends in Biotechnology, 2018, 36(1): 30-44. doi: 10.1016/j.tibtech.2017.09.003 [15] LI S N, SHOW P L, NGO H H, et al. Algae-mediated antibiotic wastewater treatment: A critical review [J]. Environmental Science and Ecotechnology, 2022, 9: 100145. doi: 10.1016/j.ese.2022.100145 [16] XIONG Q, HU L X, LIU Y S, et al. Microalgae-based technology for antibiotics removal: From mechanisms to application of innovational hybrid systems [J]. Environment International, 2021, 155: 106594. doi: 10.1016/j.envint.2021.106594 [17] XIONG J Q, KURADE M B, KIM J R, et al. Ciprofloxacin toxicity and its co-metabolic removal by a freshwater microalga Chlamydomonas mexicana [J]. Journal of Hazardous Materials, 2017, 323: 212-219. doi: 10.1016/j.jhazmat.2016.04.073 [18] XIONG Q, LIU Y S, HU L X, et al. Co-metabolism of sulfamethoxazole by a freshwater microalga Chlorella pyrenoidosa [J]. Water Research, 2020, 175: 115656. doi: 10.1016/j.watres.2020.115656 [19] RAMANAN R, KIM B H, CHO D H, et al. Algae-bacteria interactions: Evolution, ecology and emerging applications [J]. Biotechnology Advances, 2016, 34(1): 14-29. doi: 10.1016/j.biotechadv.2015.12.003 [20] ARBIB Z, de GODOS I, RUIZ J, et al. Optimization of pilot high rate algal ponds for simultaneous nutrient removal and lipids production [J]. Science of the Total Environment, 2017, 589: 66-72. doi: 10.1016/j.scitotenv.2017.02.206 [21] MENNAA F Z, ARBIB Z, PERALES J A. Urban wastewater photobiotreatment with microalgae in a continuously operated photobioreactor: Growth, nutrient removal kinetics and biomass coagulation-flocculation [J]. Environmental Technology, 2019, 40(3): 342-355. doi: 10.1080/09593330.2017.1393011 [22] PENG F Q, YING G G, YANG B, et al. Biotransformation of the flame retardant tetrabromobisphenol-A (TBBPA) by freshwater microalgae [J]. Environmental Toxicology and Chemistry, 2014, 33(8): 1705-1711. doi: 10.1002/etc.2589 [23] PENG F Q, YING G G, YANG B, et al. Biotransformation of progesterone and norgestrel by two freshwater microalgae (Scenedesmus obliquus and Chlorella pyrenoidosa): Transformation kinetics and products identification [J]. Chemosphere, 2014, 95: 581-588. doi: 10.1016/j.chemosphere.2013.10.013 [24] WANG Y, GONG X Y, HUANG D Y, et al. Increasing oxytetracycline and enrofloxacin concentrations on the algal growth and sewage purification performance of an algal-bacterial consortia system [J]. Chemosphere, 2022, 286: 131917. doi: 10.1016/j.chemosphere.2021.131917 [25] ZHANG B, LI W, GUO Y, et al. Microalgal-bacterial consortia: From interspecies interactions to biotechnological applications [J]. Renewable and Sustainable Energy Reviews, 2020, 118: 109563. doi: 10.1016/j.rser.2019.109563 [26] XIAO G X, CHEN J Q, SHOW P L, et al. Evaluating the application of antibiotic treatment using algae-algae/activated sludge system [J]. Chemosphere, 2021, 282: 130966. doi: 10.1016/j.chemosphere.2021.130966 [27] LIU Y H, WANG Z Z, YAN K, et al. A new disposal method for systematically processing of ceftazidime: The intimate coupling UV/algae-algae treatment [J]. Chemical Engineering Journal, 2017, 314: 152-159. doi: 10.1016/j.cej.2016.12.110 [28] VILLAR-NAVARRO E, BAENA-NOGUERAS R M, PANIW M, et al. Removal of pharmaceuticals in urban wastewater: High rate algae pond (HRAP) based technologies as an alternative to activated sludge based processes [J]. Water Research, 2018, 139: 19-29. doi: 10.1016/j.watres.2018.03.072 [29] HOM-DIAZ A, JAÉN-GIL A, BELLO-LASERNA I, et al. Performance of a microalgal photobioreactor treating toilet wastewater: Pharmaceutically active compound removal and biomass harvesting [J]. Science of the Total Environment, 2017, 592: 1-11. doi: 10.1016/j.scitotenv.2017.02.224 [30] JAÉN-GIL A, HOM-DIAZ A, LLORCA M, et al. An automated on-line turbulent flow liquid-chromatography technology coupled to a high resolution mass spectrometer LTQ-Orbitrap for suspect screening of antibiotic transformation products during microalgae wastewater treatment [J]. Journal of Chromatography A, 2018, 1568: 57-68. doi: 10.1016/j.chroma.2018.06.027 [31] PROSENC F, PIECHOCKA J, ŠKUFCA D, et al. Microalgae-based removal of contaminants of emerging concern: Mechanisms in Chlorella vulgaris and mixed algal-bacterial cultures [J]. Journal of Hazardous Materials, 2021, 418: 126284. doi: 10.1016/j.jhazmat.2021.126284 [32] LÓPEZ-SERNA R, GARCÍA D, BOLADO S, et al. Photobioreactors based on microalgae-bacteria and purple phototrophic bacteria consortia: A promising technology to reduce the load of veterinary drugs from piggery wastewater [J]. Science of the Total Environment, 2019, 692: 259-266. doi: 10.1016/j.scitotenv.2019.07.126 [33] ZAMBRANO J, GARCíA-ENCINA P A, HERNÁNDEZ F, et al. Removal of a mixture of veterinary medicinal products by adsorption onto a Scenedesmus almeriensis microalgae-bacteria consortium [J]. Journal of Water Process Engineering, 2021, 43: 102226. doi: 10.1016/j.jwpe.2021.102226 [34] da SILVA RODRIGUES D A, da CUNHA C C R F, FREITAS M G, et al. Biodegradation of sulfamethoxazole by microalgae-bacteria consortium in wastewater treatment plant effluents [J]. Science of the Total Environment, 2020, 749: 141441. doi: 10.1016/j.scitotenv.2020.141441 [35] XIE B H, TANG X B, NG H Y, et al. Biological sulfamethoxazole degradation along with anaerobically digested centrate treatment by immobilized microalgal-bacterial consortium: Performance, mechanism and shifts in bacterial and microalgal communities [J]. Chemical Engineering Journal, 2020, 388: 124217. doi: 10.1016/j.cej.2020.124217 [36] GARCÍA-GALÁN M J, ARASHIRO L, SANTOS L H M L M, et al. Fate of priority pharmaceuticals and their main metabolites and transformation products in microalgae-based wastewater treatment systems [J]. Journal of Hazardous Materials, 2020, 390: 121771. doi: 10.1016/j.jhazmat.2019.121771 [37] NORVILL Z N, TOLEDO-CERVANTES A, BLANCO S, et al. Photodegradation and sorption govern tetracycline removal during wastewater treatment in algal ponds [J]. Bioresource Technology, 2017, 232: 35-43. doi: 10.1016/j.biortech.2017.02.011 [38] de GODOS I, MUÑOZ R, GUIEYSSE B. Tetracycline removal during wastewater treatment in high-rate algal ponds [J]. Journal of Hazardous Materials, 2012, 229/230: 446-449. doi: 10.1016/j.jhazmat.2012.05.106 [39] HOM-DIAZ A, NORVILL Z N, BLÁNQUEZ P, et al. Ciprofloxacin removal during secondary domestic wastewater treatment in high rate algal ponds [J]. Chemosphere, 2017, 180: 33-41. doi: 10.1016/j.chemosphere.2017.03.125 [40] da SILVA RODRIGUES D A, da CUNHA C C R F, DO ESPIRITO SANTO D R, et al. Removal of cephalexin and erythromycin antibiotics, and their resistance genes, by microalgae-bacteria consortium from wastewater treatment plant secondary effluents [J]. Environmental Science and Pollution Research International, 2021, 28(47): 67822-67832. doi: 10.1007/s11356-021-15351-x [41] YU Y, ZHOU Y Y, WANG Z L, et al. Investigation of the removal mechanism of antibiotic ceftazidime by green algae and subsequent microbic impact assessment [J]. Scientific Reports, 2017, 7(1): 4168. doi: 10.1038/s41598-017-04128-3 [42] NGUYEN H T, YOON Y, NGO H H, et al. The application of microalgae in removing organic micropollutants in wastewater [J]. Critical Reviews in Environmental Science and Technology, 2021, 51(12): 1187-1220. doi: 10.1080/10643389.2020.1753633 [43] KIKI C, RASHID A, WANG Y W, et al. Dissipation of antibiotics by microalgae: Kinetics, identification of transformation products and pathways [J]. Journal of Hazardous Materials, 2020, 387: 121985. doi: 10.1016/j.jhazmat.2019.121985 [44] BAI X L, ACHARYA K. Removal of trimethoprim, sulfamethoxazole, and triclosan by the green alga Nannochloris sp. [J]. Journal of Hazardous Materials, 2016, 315: 70-75. doi: 10.1016/j.jhazmat.2016.04.067 [45] LIU Y, WANG F, CHEN X, et al. Cellular responses and biodegradation of amoxicillin in Microcystis aeruginosa at different nitrogen levels [J]. Ecotoxicology and Environmental Safety, 2015, 111: 138-145. doi: 10.1016/j.ecoenv.2014.10.011 [46] XIONG J Q, KURADE M B, JEON B H. Biodegradation of levofloxacin by an acclimated freshwater microalga, Chlorella vulgaris [J]. Chemical Engineering Journal, 2017, 313: 1251-1257. doi: 10.1016/j.cej.2016.11.017 [47] BEN W W, QIANG Z M, YIN X W, et al. Adsorption behavior of sulfamethazine in an activated sludge process treating swine wastewater [J]. Journal of Environmental Sciences, 2014, 26(8): 1623-1629. doi: 10.1016/j.jes.2014.06.002 [48] OLIVEIRA G H D, SANTOS-NETO A J, ZAIAT M. Evaluation of sulfamethazine sorption and biodegradation by anaerobic granular sludge using batch experiments [J]. Bioprocess and Biosystems Engineering, 2016, 39(1): 115-124. doi: 10.1007/s00449-015-1495-3 [49] SARMAH A K, MEYER M T, BOXALL A B A. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment [J]. Chemosphere, 2006, 65(5): 725-759. doi: 10.1016/j.chemosphere.2006.03.026 [50] YANG S F, LIN C F, YU-CHEN LIN A, et al. Sorption and biodegradation of sulfonamide antibiotics by activated sludge: Experimental assessment using batch data obtained under aerobic conditions [J]. Water Research, 2011, 45(11): 3389-3397. doi: 10.1016/j.watres.2011.03.052 [51] DANESHVAR E, ZARRINMEHR M J, HASHTJIN A M, et al. Versatile applications of freshwater and marine water microalgae in dairy wastewater treatment, lipid extraction and tetracycline biosorption [J]. Bioresource Technology, 2018, 268: 523-530. doi: 10.1016/j.biortech.2018.08.032 [52] PI S S, LI A, WEI W, et al. Synthesis of a novel magnetic nano-scale biosorbent using extracellular polymeric substances from Klebsiella sp. J1 for tetracycline adsorption [J]. Bioresource Technology, 2017, 245: 471-476. doi: 10.1016/j.biortech.2017.08.190 [53] 吴科比, 周进, 蔡中华. 藻际环境微生态结构与功能的研究进展 [J]. 生命科学, 2021, 33(5): 535-545. WU K B, ZHOU J, CAI Z H. Review of algal phycosphere: Structure and ecological function [J]. Chinese Bulletin of Life Sciences, 2021, 33(5): 535-545(in Chinese).
[54] SHENG G P, YU H Q, LI X Y. Extracellular polymeric substances (EPS) of microbial aggregates in biological wastewater treatment systems: A review [J]. Biotechnology Advances, 2010, 28(6): 882-894. doi: 10.1016/j.biotechadv.2010.08.001 [55] XIAO R, ZHENG Y. Overview of microalgal extracellular polymeric substances (EPS) and their applications [J]. Biotechnology Advances, 2016, 34(7): 1225-1244. doi: 10.1016/j.biotechadv.2016.08.004 [56] WANG L F, LI Y, WANG L, et al. Responses of biofilm microorganisms from moving bed biofilm reactor to antibiotics exposure: Protective role of extracellular polymeric substances [J]. Bioresource Technology, 2018, 254: 268-277. doi: 10.1016/j.biortech.2018.01.063 [57] ZHANG H Q, JIA Y Y, KHANAL S K, et al. Understanding the role of extracellular polymeric substances on ciprofloxacin adsorption in aerobic sludge, anaerobic sludge, and sulfate-reducing bacteria sludge systems [J]. Environmental Science & Technology, 2018, 52(11): 6476-6486. [58] SONG C, SUN X F, XING S F, et al. Characterization of the interactions between tetracycline antibiotics and microbial extracellular polymeric substances with spectroscopic approaches [J]. Environmental Science and Pollution Research, 2014, 21(3): 1786-1795. doi: 10.1007/s11356-013-2070-6 [59] SUN M, LIN H, GUO W, et al. Bioaccumulation and biodegradation of sulfamethazine in Chlorella pyrenoidosa [J]. Journal of Ocean University of China, 2017, 16(6): 1167-1174. doi: 10.1007/s11802-017-3367-8 [60] XIONG J Q, KURADE M B, JEON B H. Ecotoxicological effects of enrofloxacin and its removal by monoculture of microalgal species and their consortium [J]. Environmental Pollution, 2017, 226: 486-493. doi: 10.1016/j.envpol.2017.04.044 [61] HENA S, GUTIERREZ L, CROUÉ J P. Removal of pharmaceutical and personal care products (PPCPs) from wastewater using microalgae: A review [J]. Journal of Hazardous Materials, 2021, 403: 124041. doi: 10.1016/j.jhazmat.2020.124041 [62] BAI X L, ACHARYA K. Algae-mediated removal of selected pharmaceutical and personal care products (PPCPs) from Lake Mead water [J]. Science of the Total Environment, 2017, 581/582: 734-740. doi: 10.1016/j.scitotenv.2016.12.192 [63] GE L, DENG H. Degradation of two fluoroquinolone antibiotics photoinduced by Fe(III)-microalgae suspension in an aqueous solution [J]. Photochemical & Photobiological Sciences, 2015, 14(4): 693-699. [64] KUMAR M S, KABRA A N, MIN B, et al. Insecticides induced biochemical changes in freshwater microalga Chlamydomonas mexicana [J]. Environmental Science and Pollution Research International, 2016, 23(2): 1091-1099. doi: 10.1007/s11356-015-4681-6 [65] FU X G, WANG H J, BAI Y, et al. Systematic degradation mechanism and pathways analysis of the immobilized bacteria: Permeability and biodegradation, kinetic and molecular simulation [J]. Environmental Science and Ecotechnology, 2020, 2: 100028. doi: 10.1016/j.ese.2020.100028 [66] XIE P, HO S H, PENG J, et al. Dual purpose microalgae-based biorefinery for treating pharmaceuticals and personal care products (PPCPs) residues and biodiesel production [J]. Science of the Total Environment, 2019, 688: 253-261. doi: 10.1016/j.scitotenv.2019.06.062 [67] XIE P, CHEN C, ZHANG C F, et al. Revealing the role of adsorption in ciprofloxacin and sulfadiazine elimination routes in microalgae [J]. Water Research, 2020, 172: 115475. doi: 10.1016/j.watres.2020.115475 [68] PENG J, HE Y Y, ZHANG Z Y, et al. Removal of levofloxacin by an oleaginous microalgae Chromochloris zofingiensis in the heterotrophic mode of cultivation: Removal performance and mechanism [J]. Journal of Hazardous Materials, 2022, 425: 128036. doi: 10.1016/j.jhazmat.2021.128036 [69] FUENTES J L, GARBAYO I, CUARESMA M, et al. Impact of microalgae-bacteria interactions on the production of algal biomass and associated compounds [J]. Marine Drugs, 2016, 14(5): 100. doi: 10.3390/md14050100 [70] BUCHAN A, LECLEIR G R, GULVIK C A, et al. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms [J]. Nature Reviews Microbiology, 2014, 12(10): 686-698. doi: 10.1038/nrmicro3326 [71] 翟春梅, 刘常宏, 吕路. 铜绿微囊藻与藻际细菌Ma-B1菌株的相互作用 [J]. 环境科学研究, 2014, 27(7): 704-710. ZHAI C M, LIU C H, LYU L. Interaction between Microcystis aeruginosa and bacterium Ma-B1 strain within phycosphere [J]. Research of Environmental Sciences, 2014, 27(7): 704-710(in Chinese).
[72] VILLA J A, RAY E E, BARNEY B M. Azotobacter vinelandii siderophore can provide nitrogen to support the culture of the green algae Neochloris oleoabundans and Scenedesmus sp. BA032 [J]. FEMS Microbiology Letters, 2014, 351(1): 70-77. doi: 10.1111/1574-6968.12347 [73] GUERRA-RENTERIA A S, GARCÍA-RAMÍREZ M A, GÓMEZ-HERMOSILLO C, et al. Metabolic pathway analysis of nitrogen and phosphorus uptake by the consortium between C. vulgaris and P. aeruginosa [J]. International Journal of Molecular Sciences, 2019, 20(8): 1978. doi: 10.3390/ijms20081978 [74] LI S N, ZHANG C F, LI F H, et al. Recent advances of algae-bacteria consortia in aquatic remediation[J]. Critical Reviews in Environmental Science and Technology, 2022: 1-25. [75] HE X Y, WANG J P, ABDOLI L, et al. Mg2+/Ca2+ promotes the adhesion of marine bacteria and algae and enhances following biofilm formation in artificial seawater [J]. Colloids and Surfaces B:Biointerfaces, 2016, 146: 289-295. doi: 10.1016/j.colsurfb.2016.06.029 [76] FLEMMING H C, WINGENDER J. The biofilm matrix [J]. Nature Reviews Microbiology, 2010, 8(9): 623-633. doi: 10.1038/nrmicro2415 [77] RAMANAN R, KANG Z, KIM B H, et al. Phycosphere bacterial diversity in green algae reveals an apparent similarity across habitats [J]. Algal Research, 2015, 8: 140-144. doi: 10.1016/j.algal.2015.02.003 [78] YOU X Q, XU N, YANG X, et al. Pollutants affect algae-bacteria interactions: A critical review [J]. Environmental Pollution, 2021, 276: 116723. doi: 10.1016/j.envpol.2021.116723 [79] MUKHERJEE S, BASSLER B L. Bacterial quorum sensing in complex and dynamically changing environments [J]. Nature Reviews Microbiology, 2019, 17(6): 371-382. doi: 10.1038/s41579-019-0186-5 [80] PRESCOTT R D, DECHO A W. Flexibility and adaptability of quorum sensing in nature [J]. Trends in Microbiology, 2020, 28(6): 436-444. doi: 10.1016/j.tim.2019.12.004 [81] CHEN X Y, HU Z, QI Y, et al. The interactions of algae-activated sludge symbiotic system and its effects on wastewater treatment and lipid accumulation [J]. Bioresource Technology, 2019, 292: 122017. doi: 10.1016/j.biortech.2019.122017 [82] LI X J, CAI F S, LUAN T G, et al. Pyrene metabolites by bacterium enhancing cell division of green alga Selenastrum capricornutum [J]. Science of the Total Environment, 2019, 689: 287-294. doi: 10.1016/j.scitotenv.2019.06.162 [83] 周真真. 小球藻促生菌的促藻生长机制与应用[D]. 北京: 中国科学院大学(中国科学院过程工程研究所), 2021. ZHOU Z Z. The mechanism and application of the consortia of Chlorella sorokiniana and its growth-promotion bacteria[D]. Beijing: Institute of Process Engineering. Chinese Academy of Sciences. 2021(in Chinese).
[84] WANG B, YAO M M, ZHOU J, et al. Growth and toxin production of Gambierdiscus spp. can be regulated by quorum-sensing bacteria [J]. Toxins, 2018, 10(7): 257. doi: 10.3390/toxins10070257 [85] ZHANG B, GUO Y, LENS P N L, et al. Effect of light intensity on the characteristics of algal-bacterial granular sludge and the role of N-acyl-homoserine lactone in the granulation [J]. Science of the Total Environment, 2019, 659: 372-383. doi: 10.1016/j.scitotenv.2018.12.250 [86] van OOSTENDE N, MOERDIJK-POORTVLIET T C W, BOSCHKER H T S, et al. Release of dissolved carbohydrates by Emiliania huxleyi and formation of transparent exopolymer particles depend on algal life cycle and bacterial activity [J]. Environmental Microbiology, 2013, 15(5): 1514-1531. doi: 10.1111/j.1462-2920.2012.02873.x [87] JI X Y, JIANG M Q, ZHANG J B, et al. The interactions of algae-bacteria symbiotic system and its effects on nutrients removal from synthetic wastewater [J]. Bioresource Technology, 2018, 247: 44-50. doi: 10.1016/j.biortech.2017.09.074 [88] MITSUTANI A, YAMASAKI I, KITAGUCHI H, et al. Analysis of algicidal proteins of a diatom-lytic marine bacterium Pseudoalteromonas sp. strain A25 by two-dimensional electrophoresis [J]. Phycologia, 2001, 40(3): 286-291. doi: 10.2216/i0031-8884-40-3-286.1 [89] MANEFIELD M, RASMUSSEN T B, HENZTER M, et al. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover[J]. Microbiology (Reading, England), 2002, 148(Pt 4): 1119-1127. [90] DOBRETSOV S, TEPLITSKI M, ALAGELY A, et al. Malyngolide from the cyanobacterium Lyngbya majuscula interferes with quorum sensing circuitry [J]. Environmental Microbiology Reports, 2010, 2(6): 739-744. doi: 10.1111/j.1758-2229.2010.00169.x [91] SAWADA I, MASEDA H, NAKAE T, et al. A quorum-sensing autoinducer enhances the mexAB-oprM efflux-pump expression without the MexR-mediated regulation in Pseudomonas aeruginosa [J]. Microbiology and Immunology, 2004, 48(5): 435-439. doi: 10.1111/j.1348-0421.2004.tb03533.x [92] KWON D H, LU C D. Polyamines increase antibiotic susceptibility in Pseudomonas aeruginosa [J]. Antimicrobial Agents and Chemotherapy, 2006, 50(5): 1623-1627. doi: 10.1128/AAC.50.5.1623-1627.2006 [93] ROY R, TIWARI M, DONELLI G, et al. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action [J]. Virulence, 2018, 9(1): 522-554. doi: 10.1080/21505594.2017.1313372 [94] ZHAO X H, YU Z X, DING T. Quorum-sensing regulation of antimicrobial resistance in bacteria [J]. Microorganisms, 2020, 8(3): 425. doi: 10.3390/microorganisms8030425 [95] 欧剑虹, 谢志雄, 等. 水平基因转移 [J]. 遗传, 2003, 25(5): 623-627. OU J H, XIE Z X, CHEN X D, et al. Horizontal gene transfer [J]. Hereditas(Beijing), 2003, 25(5): 623-627(in Chinese).
[96] KOUZUMA A, WATANABE K. Exploring the potential of algae/bacteria interactions [J]. Current Opinion in Biotechnology, 2015, 33: 125-129. doi: 10.1016/j.copbio.2015.02.007 [97] SCHÖNKNECHT G, CHEN W H, TERNES C M, et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote [J]. Science, 2013, 339(6124): 1207-1210. doi: 10.1126/science.1231707 [98] RAYMOND J A, REMIAS D. Ice-binding proteins in a chrysophycean snow alga: Acquisition of an essential gene by horizontal gene transfer [J]. Frontiers in Microbiology, 2019, 10: 2697. doi: 10.3389/fmicb.2019.02697 [99] KEELING P J, PALMER J D. Horizontal gene transfer in eukaryotic evolution [J]. Nature Reviews Genetics, 2008, 9(8): 605-618. doi: 10.1038/nrg2386 [100] KEELING P J. Functional and ecological impacts of horizontal gene transfer in eukaryotes [J]. Current Opinion in Genetics & Development, 2009, 19(6): 613-619. [101] LUKAČIŠINOVÁ M, BOLLENBACH T. Toward a quantitative understanding of antibiotic resistance evolution [J]. Current Opinion in Biotechnology, 2017, 46: 90-97. doi: 10.1016/j.copbio.2017.02.013 [102] ZHANG Q, ZHANG Z Y, LU T, et al. Cyanobacterial blooms contribute to the diversity of antibiotic-resistance genes in aquatic ecosystems [J]. Communications Biology, 2020, 3: 737. doi: 10.1038/s42003-020-01468-1 [103] MICHELON W, da SILVA M L B, MATTHIENSEN A, et al. Microalgae produced during phycoremediation of swine wastewater contains effective bacteriostatic compounds against antibiotic-resistant bacteria [J]. Chemosphere, 2021, 283: 131268. doi: 10.1016/j.chemosphere.2021.131268 [104] LITTLE S M, SENHORINHO G N A, SALEH M, et al. Antibacterial compounds in green microalgae from extreme environments: A review [J]. ALGAE, 2021, 36(1): 61-72. doi: 10.4490/algae.2021.36.3.6 -




 下载:
下载: