-
在石油的开采、冶炼和加工中,不可避免地存在由于石油泄漏而带来的土壤污染问题. 石油烃外碳源向土壤的大量输入,引起土壤氮源相对不足是导致土壤微生物活性降低、抑制土著微生物降解石油烃的主要因素. 因此,向污染土壤中加入外氮源进行生物刺激修复可对石油烃降解起到促进作用. 许多研究对利用生物刺激剂修复石油污染土壤进行了详细报道. 所用氮源类型包括无机氮源NH4Cl、KNO3、NH4NO3、(NH4)2SO4,有机氮源尿素和有机肥等[1 − 3]. 研究认为,当土壤C/N比接近10时,土壤微生物的活性最好,对石油烃的去除能力最强[4].
土壤菌群结构多样性和功能菌群活性对于生物技术的实施至关重要. 目前多是利用变性梯度凝胶电泳(denaturing gradient gel electrophoresis, DGGE)、实时定量聚合酶链反应(quantitative real-time polymerase chain reaction, qPCR)、高通量测序(high throughput sequencing)、宏基因组学(metagenomics)等技术对土壤菌群结构和功能代谢基因表达进行研究[5 − 7]. 修复过程中可降解石油烃的功能菌属主要有细菌芽孢杆菌属(Bacillus)、铜绿假单胞菌属(Pseudomonas)、不动细菌属(Acinetobacter)、红球菌属(Rhodococcus)、原小单孢菌属(Promicromonospora),以及真菌的担子菌门(Basidiomycota)和子囊菌门(Ascomycota)等[8 − 10]. 生物刺激修复可较好地保持土壤微生物菌群结构多样性和稳定性,有利于石油烃的持续降解[11 − 12]. 现有研究阐明了石油烃降解功能菌属类别以及土壤菌群稳定性对生物修复的影响作用,但是对于不同功能类群参与石油烃降解的作用强度和利用特征尚不清楚.
磷脂脂肪酸(Phospholipid fatty acids, PLFA)是几乎所有活体细胞膜的主要成分,周转速率极快且随细胞死亡而迅速降解. 不同类群的微生物PLFA组成具有一定差异,因而磷脂脂肪酸可作为生物标识物用以描述微生物类群对不同碳源的利用特征 [13 − 14]. 目前PLFA技术主要用于研究农业上施肥、植物残体及植被种植等因素对土壤微生物群落活性的影响[15 − 17],较少用于研究污染土壤生物修复过程中功能微生物类群对石油烃组分的利用特征.
烷烃是石油烃的重要组分之一,本文以13C-十六烷标记的污染土壤为研究对象,通过向污染土壤中加入硝酸钾和有机肥进行生物刺激修复处理,利用稳定同位素标记-磷脂脂肪酸技术(13C -SIP-PLFA)探究十六烷中碳组分被不同土壤微生物类群的利用情况,本研究可为深入理解石油烃的生物降解代谢过程提供参考.
-
实验所用土壤为取自陕西省延安市安塞县未受石油污染的黄绵土. 土壤基本理化性质采用常规方法进行分析[18](表1). 土壤经剔除可见杂质、自然风干、研磨粉碎后将其铺平摊薄,去除土壤中残留的细微植物残体和根须,随后过100目筛并充分混匀后,装于自封袋中,−80 ℃下储存备用.
所用腐熟有机肥取自西北农林科技大学,是以干质量比1:2的猪粪:稻壳加入5.0%的木炭渣经堆制腐解而成. 有机肥中的主要元素含量为C、N、P、K、Na、Ca、Mg、Al、S、Cu、Fe、Mn、Mo、Zn分别为185600、20400、6500、36200、3810、2130、2420、1330、1575、400、1615、284、4.46、140 mg·kg−1.
-
13C-十六烷配制:0.2500 g丰度99% atom的标记十六烷(十六烷-1,2-13C2, Sigma公司)与4.7500 g丰度1.108% atom的分析纯十六烷(Sigma 公司)充分混合,计算其13C丰度为5.976% atom.
实验方案设计如表2所示. 取9份100 g晾干磨碎并过100目筛的土壤分3组(每组3个平行),分别加入5000 mg·kg−1 13C-十六烷标记物、5000 mg·kg-1 13C-十六烷标记物和硝酸钾(调节土壤碳氮比为10/1)、5000 mg·kg-1 13C-十六烷标记物和有机肥(添加量150 g·kg−1),将各组土壤充分混合均匀.3组处理编号分别为:CC、CN和CY.室温条件下在0、3、30 d采集各组土壤样品,于−80 ℃下保存待测.
-
土壤中正十六烷利用超声萃取提取-GC/MS测定. 超声提取和GC/MS测定方法详见文献[19].
土壤PLFA的提取 微生物群落结构用土壤磷脂脂肪酸(phospholipid fatty acid,PLFA)表示. 土壤PLFA的提取方法见文献[19]. 基本步骤为:利用氯仿:甲醇:柠檬酸=1:2:0.8作为提取剂提取土壤中的磷脂、糖脂和中性脂,以氯仿和丙酮作为洗脱剂,利用SPE柱(Supelco公司)分离去除糖脂和中性脂,向提取获得的磷脂中加入甲醇:甲苯(1:1)的混合液和0.2 mol·L−1 KOH甲醇溶液进行甲酯化后,加入蒸馏水和冰醋酸,再用正己烷萃取获得磷脂脂肪酸甲酯,用氮气吹干后,在酯化的样品中加入19:0甲基酯作内标,将磷脂脂肪酸甲酯溶解于60 μL正己烷中.
土壤PLFA碳同位素的测定 用气相色谱仪(Trace GC 1310 +GC IsoLink Ⅱ连ConFlo Ⅳ(测可气化有机物中的CNHO))进行检测,利用MIDI SHERLOCCS微生物鉴定系统进行脂肪酸的比对鉴定. 脂肪酸的定量采用峰面积和内标曲线法. 单个PLFA的13C/12C用气相色谱-燃烧法-稳定同位素比质谱仪(GC-C-IRMS)进行测定,测定方法详见文献[19].
共检测出含有13C-同位素的18种PLFA单体,以18:1ω9c指示真菌;以16:0(10Me)、17:0(10Me)、18:0(10Me)、17:1ω7c(10Me)指示放线菌;以i15:0、i16:0、i17:0、a15:0、a17:0指示革兰氏阳性菌(G+);以16:1ω5c、16:1ω7c、18:1ω5c、18:1ω7c、cy17:0ω7c、cy19:0ω7c指示革兰氏阴性菌(G−);以16:0、18:0指示Unspecific菌[20]. 将所有微生物PLFAs总和表征为总微生物量.
-
土壤中各磷脂脂肪酸单体含量为:
式中,PF为土壤中磷脂脂肪酸单体的含量,nmol·g−1;AF和ASTD为样品和内标物(甲基酯19:00)的峰面积;WSTD为内标物(甲基酯19:00)的质量,μg;W为干土重量,g;M为各磷脂脂肪酸单体的相对分子质量,g·mol−1.
稳定同位素比率计算:
式中,Rsample为样品13C/12C原子比值;Rstandard为标准品(美国卡罗莱纳州白垩纪皮狄组层位中的拟箭石化石, PDB)原子比值,数值为0.011802.
PLFA单体中13C的量:
式中,13C-PF为PLFA单体中的13C的量,ng·g−1;(%atom13C)LP和(%atom13C)ULP分别为标记样品PLFA和未标记样品PLFA单体中的%atom13C,LPC为标记样品PLFA单体中碳含量,ng·g−1.
-
采用Excel2010和SPSS 25.0进行数据正态性和方差齐性检验,同一时期不同处理、同一处理不同时期差异显著性用One-way ANOVA(单因素方差分析)检验,利用LSD, P<0.05(最小显著差数法)进行多重比较,采用Origin 2021绘制图表,数据表示为平均值±标准差.
-
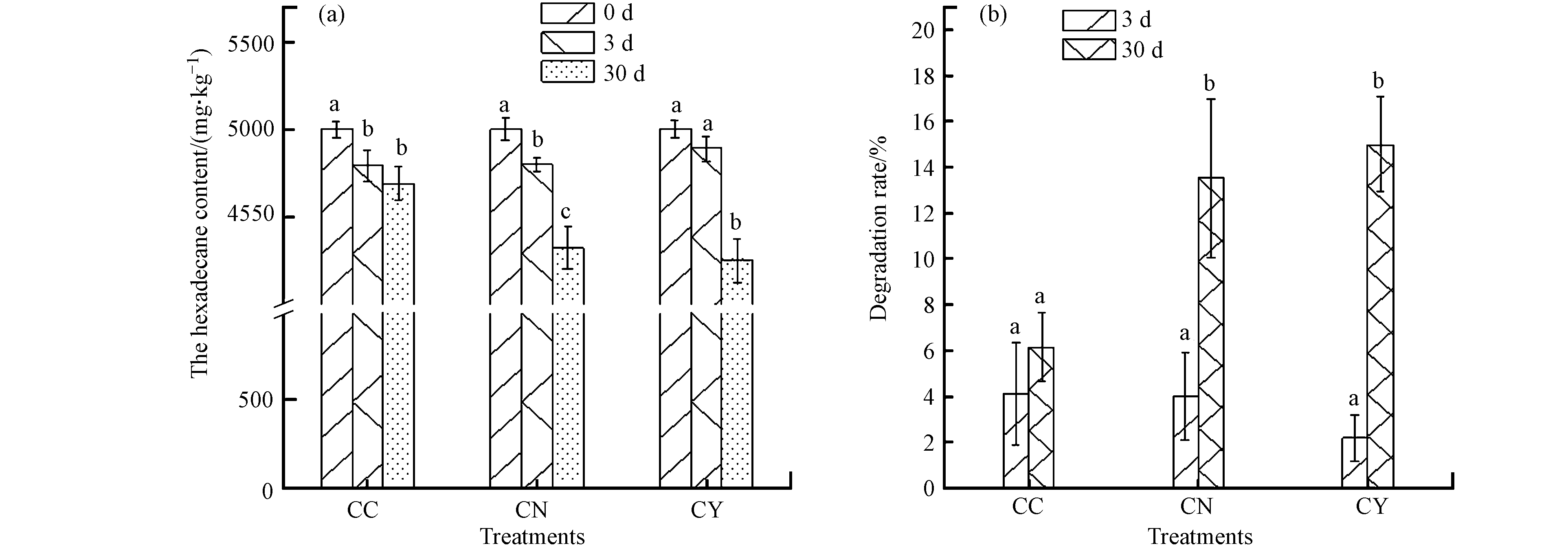
向十六烷污染土壤中分别加入KNO3、有机肥进行生物刺激修复,比较了自然降解(CC)、KNO3生物刺激修复(CN)、有机肥生物刺激修复(CY)对十六烷的去除效果(图1).
修复初期(修复3 d),十六烷在自然降解和加入KNO3修复处理的土壤中降解较为迅速,烷烃含量由5000 mg·kg−1降低至4793 mg·kg−1和4800 mg·kg−1,加入有机肥修复处理的土壤中烷烃含量为4891 mg·kg−1. 修复30 d时,加入两种修复剂处理的土壤中烷烃去除效率显著提高,十六烷含量降低至4323 mg·kg−1(CN)和4250 mg·kg−1(CY)(图1a). 与未经修复处理的土壤相比,烷烃降解率分别提高为自然条件下的2.21倍和2.44倍,说明加入外源氮进行生物刺激修复可有效提高土壤微生物对烷烃的降解效率(图1b).
-
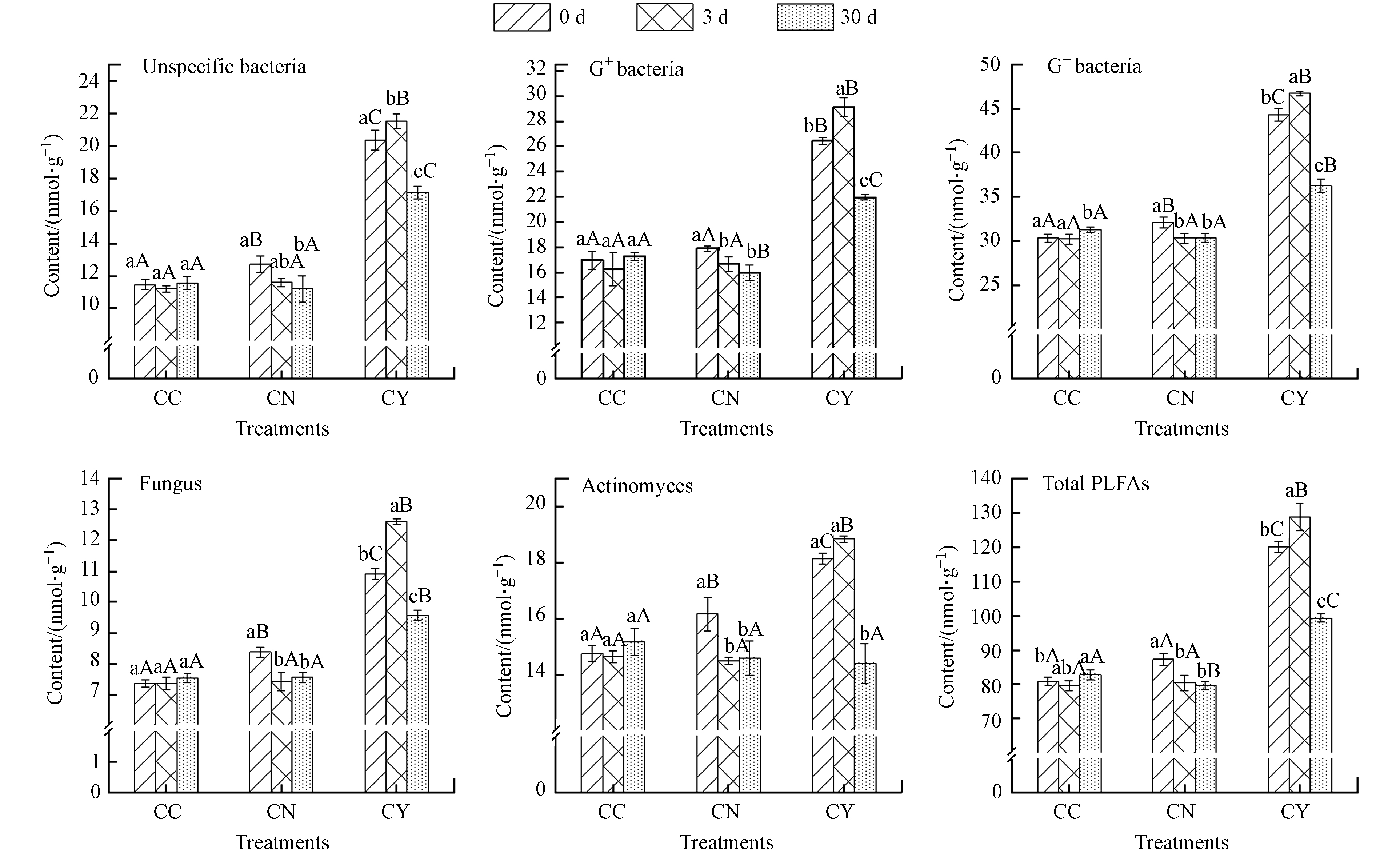
修复期间,自然降解(CC)的土壤中PLFA总量变化不明显,为80.82—82.83 nmol·g−1. 各微生物类群PLFA含量由高到低依次为:G-(30.30 nmol·g−1)> G+(16.96 nmol·g−1)>放线菌(14.76 nmol·g−1)>Unspecific 菌(11.44 nmol·g−1)>真菌(7.36 nmol·g−1). 土壤中含量较多的9种PLFA单体(大于3 nmol·g−1)包括 Unspecific菌16:00,G+菌i15:0、a15:0,G-菌16:1ω5c、16:1ω7c、18:1ω7c、cy19:0ω7c,真菌18:1ω9c和放线菌16:0(10Me)(图2, 图3),其PLFA单体的含量分别为9.42、6.55、3.96、4.91、6.45、11.05、4.11、7.36、9.70 nmol·g−1.
向污染土壤中加入KNO3修复处理3 d,PLFA总量略有降低;修复30 d 时,PLFA总量为79.70 nmol·g−1. 5种微生物类群G-、 G+、放线菌、Unspecific菌、真菌的PLFA含量分别为30.36、15.98、14.60、11.20、7.56 nmol·g−1,各微生物类群和9种PLFA单体在修复期间含量变化不大. 加入有机肥修复处理初期(修复3 d)的各微生物类群PLFA含量和总PLFA含量增加,修复至30 d时显著降低,土壤中总PLFA为99.33 nmol·g−1,5种微生物类群PLFA含量为36.26、21.97、14.41、9.57、17.12 nmol·g−1.
十六烷污染和修复处理的土壤样品中不同微生物类群特征脂肪酸含量由高到低依次为:G-细菌>G+细菌>放线菌>Unspecific菌>真菌. 与未经修复处理的土壤(CC)相比,加入KNO3修复处理(CN)对土壤中的总PLFA和各生物类群PLFA影响较小,有机肥施入(CY)显著增加了土壤中总PLFA含量,其中Unspecific菌、G+、G-菌PLFA的增量最大(图2).
总体而言,加入修复剂处理的G+/G−的PLFA比值随时间变化不显著,但对细菌/真菌的PLFA比值影响较显著(表3). 加入有机肥进行修复对 G + /G-的 PLFA 比值有显著影响,对细菌/真菌的的 PLFA 比值影响不显著. 加入有机肥进行修复 G + /G-比值高于自然降解和加入硝酸钾进行修复处理的土壤.加入有机肥进行修复对G +/G-的PLFA比值有显著影响,对细菌/真菌的的PLFA比值影响不显著. 加入有机肥进行修复G+ /G-比值高于自然降解和加入硝酸钾进行修复处理的土壤.
-
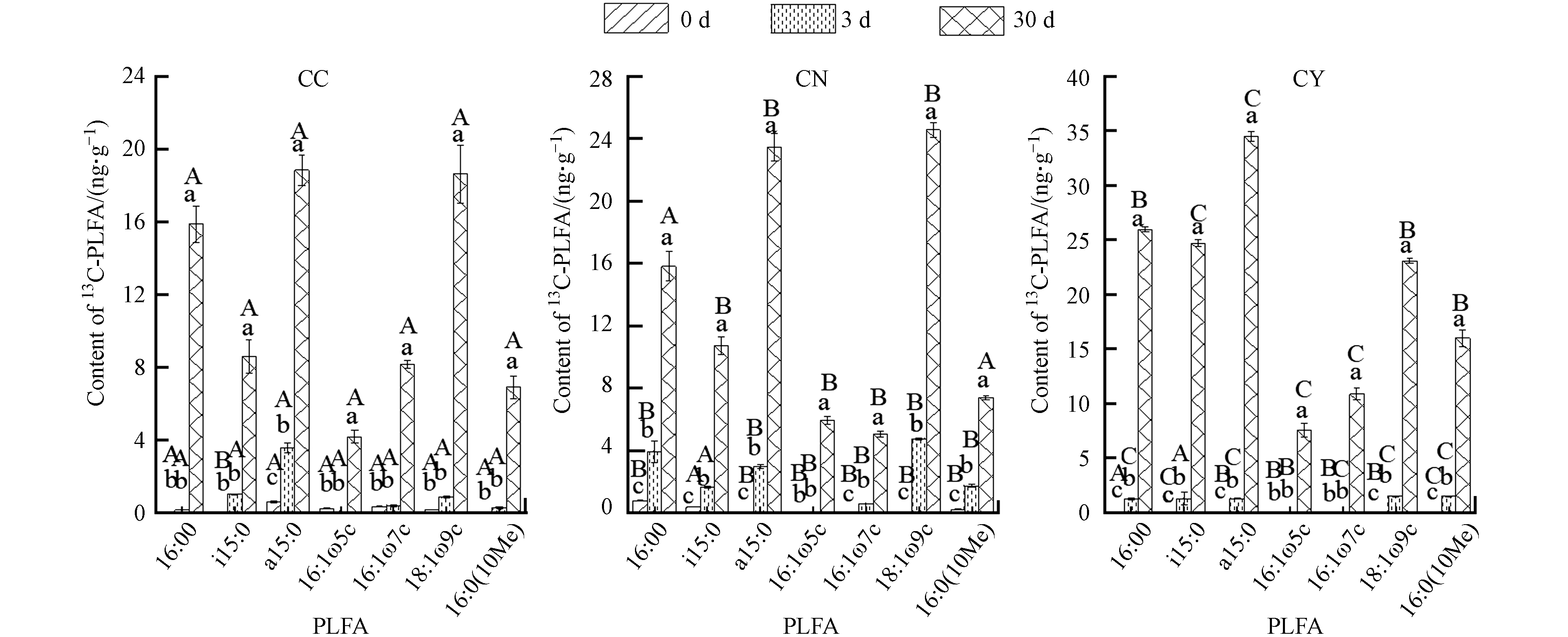
通过测定不同修复处理土壤微生物PLFA单体中13C含量可以了解各微生物类群对十六烷的利用情况. 根据图4可知,在不同修复处理的土壤中,可利用十六烷的微生物主要有G+菌i15:0、a15:0,G-菌16:1ω5c、16:1ω7c,真菌18:1ω9c、放线菌16:0(10Me)和Unspecific菌16:00.土壤中烷烃的降解代谢需要5种不同微生物类群的协同作用. 在3种修复处理进行至3 d时,各微生物类群PLFA中13C含量较低. 修复至30 d时,3种修复处理的土壤微生物类群中的13C含量显著增加,说明修复30 d时土壤微生物已适应了十六烷的污染,并以十六烷作为碳源底物进行同化代谢.
自然降解30 d的土壤(CC)中,i15:0、a15:0、16:1ω5c、16:1ω7c 、18:1ω9c、16:0(10Me)和16:007种PLFA单体中的13C总量为81.12 ng·g−1,其中G+(i15:0、a15:0)和真菌(18:1ω9c)中13C含量最高,分别为27.41 ng·g−1和18.61 ng·g−1,占7种PLFA单体中13C总量的33.8%和22.9%.
加入硝酸钾作为生物刺激剂修复30 d 时, 上述7种PLFA单体中13C总量增加至92.84 ng·g−1,5种可同化代谢十六烷的微生物类群中,G+菌(i15:0+a15:0)和真菌(18:1ω9c)PLFA中13C含量分别增加了6.72 ng·g−1和5.94 ng·g−1,Unspecific菌、G-菌、放线菌PLFA中13C含量变化不大.
加入有机肥进行修复处理30 d, 7种PLFA单体中13C总量增加至142.67 ng·g−1,5种微生物类群PLFA中13C含量均增加,其中G+菌(i15:0+a15:0)、Unspecific菌(16:00)、放线菌(6:0(10Me))PLFA中的13C含量增加最明显,增量分别为31.77、10.19、9.11 ng·g−1. 说明向十六烷污染土壤中加入硝酸钾或者有机肥进行修复处理,可以提高土壤微生物类群对十六烷的同化利用,但是两种修复剂对土壤中不同微生物类群代谢十六烷的激活作用上存在差异,加入有机肥修复处理对微生物同化代谢污染物的激活作用更为显著.
图5为不同生物类群PLFA单体中的13C含量,图6为不同处理土壤中各类群PLFA利用13C的相对丰度. 根据图5和图6,在加入硝酸钾修复的土壤中,各微生物类群PLFA中13C含量由高到低顺序为:G+菌>真菌> Unspecific 菌> G-菌>放线菌;加入有机肥修复的土壤中各微生物类群的13C含量由高到低的顺序为:G+菌>Unspecific菌>真菌> G-菌>放线菌. 总体上,两种修复处理的土壤中革兰氏阳性菌对十六烷的利用率最高,放线菌对十六烷的利用率最低. 加入有机肥修复处理可显著促进G+菌和Unspecific菌对十六烷的同化代谢.
不同类群微生物的磷脂脂肪酸(PLFA)组成具有一定差异,因此PLFA可用于表征土壤中各微生物类群的活性.PLFA单体中的碳主要来源于土壤有机质以及添加的外源碳(该文中为13C-十六烷中的碳),因此可利用13C -SIP-PLFA技术研究可利用十六烷进行同化代谢作用的微生物群落变化情况. 土壤中外碳源的大量引入导致了土壤中氮素的相对不足. 在对石油烃污染土壤进行修复时,根据石油烃中碳素含量(一般按石油烃含量的85%计),加入硝酸钾等外源氮调节土壤C/N比(一般为10/1)是常用的污染土壤修复方式. 有研究认为外源氮的加入可提高土壤中石油烃降解功能菌的活性[21],也有研究认为在加入硝态氮修复柴油污染土壤时,从亚硝酸盐到铵的转化受到低丰度亚硝酸盐还原酶的限制,进而降低了了石油烃的降解速率[22]. 本文利用硝酸钾作为生物刺激剂对污染土壤修复30 d,可有效提高土壤中十六烷的去除效率.PLFA分析结果表明,加入硝酸钾进行生物刺激修复使得土壤中活性微生物数量略有降低(土壤PLFA总量由82.8 nmol·g−1降低至79.7 nmol·g−1),但是PLFA中的13C含量、尤其是G+菌和真菌PLFA中13C含量明显增加,说明在加入硝酸钾修复的土壤中,尽管烷烃降解功能类群对十六烷同化代谢作用的增强与烷烃降解效率存在一定的相关关系,但土壤中总微生物活性并不能指示功能类群对十六烷的同化代谢作用.
有机肥中含有丰富的微生物和养分. 农业上向种植作物的土壤中施入有机肥能改善作物根际微生物菌群,提高植物的抗病虫能力[23]. 在环境领域,用有机肥作为生物刺激剂修复污染土壤,可促进石油烃污染物的降解[24 − 25]. 本文中,有机肥的施入显著提高了土壤微生物的总PLFA含量,并且各微生物类群PLFA中的13C含量也明显增加,说明有机肥的施入同时提高了土壤总微生物和烷烃代谢功能类群的活性. 这与施入硝酸钾修复对土壤总微生物活性的影响作用存在差异. 多数农田中革兰氏阳性菌数量多于阴性菌[16]. 本文中3种修复处理土壤中G+/G−的PLFA比值约为0.55—0.60,可能是由于重度烃污染改变了土壤中G+/G−微生物的类群分布,使得土壤中革兰氏阴性菌数量多于阳性菌. 一些文献报道了在石油烃污染的土壤中,大量的革兰氏阴性菌(主要是变形菌)得以富集[26 − 27],论文的研究结果和文献报道一致. Atlas和Bartha[28]利用r-K繁殖策略对土壤受到石油烃污染后革兰氏阴性菌的富集进行解释,认为革兰氏阴性菌多是r-繁殖策略者,增殖速率快且种群增长率大,因此能够在重度污染的土壤环境中短时间内快速繁殖.
尽管在重度烃污染的土壤中富集生长了大量的革兰氏阴性菌,然而,污染土壤中革兰氏阳性菌的13C-PLFA含量却高于革兰氏阴性菌,革兰氏阳性菌是对十六烷同化代谢能力最强的微生物类群. 加入两种不同生物刺激剂进行修复处理时,G+菌PLFA中的13C增量最多,说明生物刺激修复对G+菌同化代谢十六烷的激活效应最强. 可能是由于G+菌具有较厚而致密的肽聚糖层(20—80 nm),对于外部环境变化具有更强的适应特性,使得许多革兰氏阳性菌在污染环境条件下产生的芽孢对于生物刺激剂具有更好的应激效应[29 − 30]. 另一方面,加入硝酸钾进行修复处理使得G+菌和真菌PLFA中13C含量增加显著(增量为6.72 ng·g−1和5.94 ng·g−1),而加入有机肥修复处理显著增加了G+菌、Unspecific 菌、放线菌PLFA中的13C含量(增量分别为31.77、10.19、9.11 ng·g−1),两种修复处理对烷烃代谢功能类群的激活效应存在差异,进一步说明两种修复剂对土壤中十六烷烃同化代谢菌的激活机制上存在差异.
根据文献报道,有机肥对土壤中有机污染物的去除作用,一方面是由于堆肥的保水、保温能力及堆肥中丰富的营养可提高土著微生物的活性. 另一方面,堆肥中的腐殖质可以降低土壤有机质对有机污染物的吸附锁定作用. 此外,有机肥中蕴含的微生物与土壤中土著微生物存在一定的作用关系(种间共处或者互生),进而促进了十六烷的降解[31].
课题组的前期研究表明,加入有机肥对石油污染土壤进行修复处理,土壤微生物多样性增加,土著优势菌属的生长则受到抑制,非优势菌的生长得以促进,同时出现了一些新的降解菌属[31]. 本文中,加入有机肥修复的土壤中微生物PLFA含量和13C-PLFA含量显著增加,说明有机肥的加入促进了土壤微生物活性和对烷烃的代谢,有机肥中的微生物和土壤土著微生物之间可能存在协同作用关系.
-
十六烷污染的土壤中,各微生物类群PLFA含量由高到低依次为:G-菌>G+菌> 放线菌>Unspecific 菌>真菌. 向十六烷重度污染的土壤中加入硝酸钾和有机肥均能促进十六烷的降解. 加入KNO3修复处理降低了总PLFA含量,但是使得G+菌和真菌PLFA中13C含量增加显著. 施入有机肥可显著增加土壤中总PLFA含量,同时使得G+菌和放线菌PLFA中的13C含量增加显著. 土壤总微生物活性并不能指示烷烃的生物降解效率,烷烃降解功能类群对烷烃同化代谢作用的增强与石油烃的降解效率存在一定的相关关系. 不同生物刺激剂对土壤中的十六烷烃同化代谢菌的激活机制上存在差异.
硝酸钾和有机肥添加对土壤中十六烷的降解效果及微生物类群活性变化研究
Effects of potassium nitrate and organic fertilizer on microbial activity in hexadecane-contaminated soil
-
摘要: 向污染土壤中加入外源氮进行生物刺激修复是目前广为采用的土壤有机污染修复技术. 然而,目前对于修复过程中土壤微生物类群对组分烃的代谢特征尚不清楚. 本文以13C标记的十六烷污染土壤为研究对象,利用稳定同位素标记-磷脂脂肪酸技术(stable isotope labeling-phospholipid fatty acid technology,13C -SIP-PLFA)研究了加入硝酸钾和有机肥对石油污染土壤进行修复时,不同微生物类群对十六烷的利用特征. 结果表明,与自然降解(CC)相比,加入KNO3(CN)和施入有机肥(CY)的处理均可提高土壤中十六烷的去除效率. 修复30 d时,土壤中十六烷的去除率由6.14% (CC)提高至13.6% (CN) 和15.0% (CY). 加入硝酸钾修复使得土壤微生物总量略有降低(总PLFAs由82.8 nmol·g−1(CC)降低至79.7 nmol·g−1(CN)),但被微生物同化为细胞组分的十六烷含量(13C-PLFA)由81.12 ng·g−1(CC)增加至92.84 ng·g−1(CN),硝酸钾生物刺激修复提高了革兰氏阳性菌和真菌对十六烷的同化代谢作用. 加入有机肥修复的土壤中,微生物总量(总PLFAs为99.3 nmol·g−1)和微生物同化代谢十六烷的含量均明显增加(13C-PLFA 为142.67 ng·g−1),土壤中革兰氏阳性菌和放线菌对十六烷的同化代谢作用明显增强. 在不同修复处理的土壤中,可利用十六烷的主要微生物有G+菌i15:0和a15:0、G-菌16:1ω5c和16:1ω7c、真菌18:1ω9c、放线菌16:0(10Me)和Unspecific菌16:00. 结果表明,革兰氏阳性菌是不同修复处理中最主要的十六烷降解菌,两种修复剂对土壤不同微生物类群的代谢激活作用存在差异.Abstract: Biostimulation by adding exogenous nitrogen to the soil is a common application technology for soil organic pollution. However, the metabolic characteristics of soil microbial communities towards the contaminants during the remediation are still not clear. In this study, two stimulants, KNO3 and compost, were applied to 13C-labeled hexadecane-contaminated soil for 30 days of remediation. The removal rates of hexadecane were determined using GC-MS, and the phospholipid fatty acid ( PLFA) contents and the 13C incorporated into PLFA was quantified using 13C -PLFA-SIP technique.Results showed that the removal rates of hexadecane was higher in the KNO3(CN)and compost (CY) amendment soils than that in the natural attenuation(CC). After 30 days of incubations, the removal rates of hexadecane enhanced from 6.14% (CC) to 13.6% (CN) and 15.0% (CY), respectively. Although KNO3 amendment slightly decreased the total microbial biomass (the total PLFAs changed from 82.8 nmol·g−1 in CC to 79.7 nmol·g−1 in CN), the 13C-PLFA increased from 81.12 ng·g−1 to 92.84 ng·g−1. KNO3 amendment improved the hexadecane assimilation utilization by Gram-positive bacteria and fungi. In the compost treated soil (CY), the total PLFAs and 13C-PLFA increased to 99.3 nmol·g−1 and 142.67 ng·g−1, respectively, and the assimilation utilization of hexadecane by Gram-positive bacteria and actinomycetes were significantly enhanced. In the different treatments, the microorganisms that can use hexadecane mainly included gram-positive bacteria i15:0 and a15:0, gram-negative bacteria 16:1ω5c and 16:1ω7c, fungus 18:1ω9c, actinomycetes 16:0 (10Me), and unspecified bacteria 16:00. The results indicated that gram-positive bacteria were the most dominant hydrocarbon-degrading bacteria in the different treatments, and the remediation characterictics of KNO3 or compost addition toward hydrocarbon-polluted soil were different.
-

-
图 4 不同处理下土壤PLFA单体中13C含量
Figure 4. The amount of 13C-PLFA in microbial monomers PLFA in different treatments (Different lowercase letters represent significant differences in 13C-PLFA content in different time (P<0.05); different uppercase letters represent significant differences in 13C-PLFA content in different treatments at the same period (P<0.05))
表 1 土壤基本性质
Table 1. Basic physicochemical properties of soil
pH 含水率/%
Humanity有机碳/(g·kg−1)
Organic carbon总氮/(mg·kg−1)
Total nitrogen铵氮/(mg·kg−1)
NH4+-N硝氮/(mg·kg−1)
NO3--N全硫/(mg·kg−1)
Total sulfur碳氮比
C/N8.20±0.04 0.12±0.01 17.1±1.27 400.1±12.5 4.11±0.16 4.01±0.15 120.0±8.9 42.5 表 2 实验方案设计
Table 2. Experimental scheme design
编号
Code处理
TreatmentCC 100 g洁净土壤+0.5 g 13C-十六烷 CN 100 g洁净土壤+0.5 g 13C-十六烷+0.3066 g KNO3(C/N=10/1) CY 100 g洁净土壤+ 0.5 g 13C-十六烷+15 g 有机肥(C/N=10/1) 表 3 不同修复处理土壤细菌 /真菌、革兰氏阳性菌 /革兰氏阴性菌PLFA比值的变化
Table 3. The ratios of bacteria to fungi and gram-positive to gram-negative bacteria under different treatments
处理
Treatment细菌/真菌
Bacteria/Fungi革兰氏阳性菌/革兰氏阴性菌
G+/G−0 d 3 d 30 d 0 d 3 d 30 d CC 7.97±0.02aA 7.82±0.03aA 7.97±0.01aA 0.56±0.01aA 0.54±0.03aA 0.55±0.04aA CN 7.49±0.11bA 7.89±0.04aB 7.61±0.05aAB 0.56±0.04aA 0.55±0.02aA 0.53±0.06aA CY 8.35±0.16cA 7.72±0.21aB 7.87±0.07aC 0.60±0.05bA 0.62±0.02bA 0.61±0.04bA 注:小写字母为同一时期不同修复之间差异性, 大写字母表示同一处理不同时期差异性, 字母不同表示差异显著(P<0.05).
Different lowercase letters indicated significant differences among different treatments (P<0.05); different uppercase letters represent significant differences in different remediation periods in the same treatments(P<0.05) -
[1] SAFDARI M S, KARIMINIA H R. Development of bioreactors for comparative study of natural attenuation, biostimulation, and bioaugmentation of petroleum-hydrocarbon contaminated soil[J]. Journal of Hazardous Materials, 2018, 342: 270-278. doi: 10.1016/j.jhazmat.2017.08.044 [2] NIKOLOPOULOU M, PASADAKIS N, NORF H, et al. Enhanced ex situ bioremediation of crude oil contaminated beach sand by supplementation with nutrients and rhamnolipids[J]. Marine Pollution Bulletin, 2013, 77(1/2): 37-44. [3] WU M L, WU J L, ZHANG X H, et al. Effect of bioaugmentation and biostimulation on hydrocarbon degradation and microbial community composition in petroleum-contaminated loessal soil[J]. Chemosphere, 2019, 237: 124456. doi: 10.1016/j.chemosphere.2019.124456 [4] VARJANI S, UPASANI V N. Influence of abiotic factors, natural attenuation, bioaugmentation and nutrient supplementation on bioremediation of petroleum crude contaminated agricultural soil[J]. Journal of Environmental Management, 2019, 245: 358-366. [5] BAO Y J, XU Z X, LI Y, et al. High-throughput metagenomic analysis of petroleum-contaminated soil microbiome reveals the versatility in xenobiotic aromatics metabolism[J]. Journal of Environmental Sciences, 2017, 56: 25-35. doi: 10.1016/j.jes.2016.08.022 [6] IQBAL A, MUKHERJEE M, RASHID J, et al. Development of plant-microbe phytoremediation system for petroleum hydrocarbon degradation: An insight from alkb gene expression and phytotoxicity analysis[J]. Science of the Total Environment, 2019, 671: 696-704. doi: 10.1016/j.scitotenv.2019.03.331 [7] TARTAGLIA M, SCIARRILLO, ZUZOLO D, et al. Exploring an enhanced rhizospheric phenomenon for pluricontaminated soil remediation: Insights from tripartite metatranscriptome analyses[J]. Journal of Hazardous Materials, 2022, 428: 128246. doi: 10.1016/j.jhazmat.2022.128246 [8] 吴蔓莉, 陈凯丽, 叶茜琼, 等. 堆肥-生物强化对重度石油污染土壤的修复作用[J]. 环境科学, 2017, 38(10): 4412-4419. WU M L, CHEN K L, YE X Q, et al. Remediation of petroleum-contaminated soil using a bioaugmented compost technique[J]. Environmental Science, 2017, 38(10): 4412-4419(in Chinese).
[9] LI Q Q, LI J B, JIANG L F, et al. Diversity and structure of phenanthrene degrading bacterial communities associated with fungal bioremediation in petroleum contaminated soil[J]. Journal of Hazardous Materials, 2021, 403: 123895. doi: 10.1016/j.jhazmat.2020.123895 [10] AMBUST S, DAS A J, KUMAR R. Bioremediation of petroleum contaminated soil through biosurfactant and Pseudomonas sp. SA3 amended design treatments[J]. Current Research in Microbial Sciences, 2021, 2: 100031. doi: 10.1016/j.crmicr.2021.100031 [11] LI Q, HUANG Y, WEN D, et al. Application of alkyl polyglycosides for enhanced bioremediation of petroleum hydrocarbon-contaminated soil using Sphingomonas changbaiensis and Pseudomonas stutzeri[J]. Science of the Total Environment, 2020, 719: 137456. doi: 10.1016/j.scitotenv.2020.137456 [12] 祁燕云, 吴蔓莉, 祝长成, 等. 基于高通量测序分析的生物修复石油污染土壤菌群结构变化[J]. 环境科学, 2019, 40(2): 869-875. QI Y Y, WU M L, ZHU C C, et al. Microbial community structure shift during bioremediation of petroleum contaminated soil using high throughput sequencing[J]. Environmental Science, 2019, 40(2): 869-875(in Chinese).
[13] 高士杰, 王春梅, 王鹏, 等. 多形态多水平氮添加对温带森林土壤根系呼吸和微生物呼吸的影响[J]. 环境化学, 2020, 39(6): 1568-1577. doi: 10.7524/j.issn.0254-6108.2020021202 GAO S J, WANG C M, WANG P, et al. Effects of multi-form and multi-level nitrogen addition on root respiration and microbial respiration in temperate forest soil[J]. Environmental Chemistry, 2020, 39(6): 1568-1577(in Chinese). doi: 10.7524/j.issn.0254-6108.2020021202
[14] 任慧琴, 赵念席, 陈磊, 等. 土壤微生物群落结构分析中磷脂脂肪酸法和温和碱性甲酯化法的比较及定量优化[J]. 环境化学, 2014, 33(5): 760-764. doi: 10.7524/j.issn.0254-6108.2014.05.008 REN H Q, ZHAO N X, CHEN L, et al. Comparison of phospholipid fatty acid method and mild alkaline methylesterefication method in characterizing soil microbial communities and quantitative optimization[J]. Environmental Chemistry, 2014, 33(5): 760-764(in Chinese). doi: 10.7524/j.issn.0254-6108.2014.05.008
[15] 陈展, 王效科, 尚鹤. 13CO2示踪臭氧胁迫对水稻土壤微生物的影响[J]. 环境科学, 2014, 35(10): 3911-3917. CHEN Z, WANG X K, SHANG H. Ozone effects on soil microbial community of rice investigated by 13C isotope labeling[J]. Environmental Science, 2014, 35(10): 3911-3917(in Chinese).
[16] 邓少虹, 郑小东, 毛婉琼, 等. 稻田与旱地土壤中真菌和细菌对秸秆碳的利用特征[J]. 环境科学, 2022, 43(2): 1069-1076. DENG S H, ZHENG X D, MAO W Q, et al. Characteristics of microbial utilization for crop residue-derived C in paddy and upland soils[J]. Environmental Science, 2022, 43(2): 1069-1076(in Chinese).
[17] 王伟华, 刘毅, 唐海明, 等. 长期施肥对稻田土壤微生物量、群落结构和活性的影响[J]. 环境科学, 2018, 39(1): 430-437. WANG W H, LIU Y, TANG H M, et al. Effects of long-term fertilization regimes on microbial biomass, community structure and activity in a paddy soil[J]. Environmental Science, 2018, 39(1): 430-437(in Chinese).
[18] 鲍士旦. 土壤农化分析[M]. 3版. 北京: 中国农业出版社, 2000. BAO S D. Soil and agricultural chemistry analysis[M]. 3rd edition. Beijing: Chinese Agriculture Press, 2000(in Chinese).
[19] 许殷瑞. 陕北采油区土壤微生物群落结构及对石油烃组分的利用机制[D]. 西安: 西安建筑科技大学, 2021. XU Y R. Soil microbial community structure and utilization mechanism of petroleum hydrocarbon components in oil production area of northern Shaanxi[D]. Xi'an: Xi'an University of Architecture and Technology, 2021(in Chinese).
[20] LIU L, ESTIARTE M, BENGTSON P, et al. Drought legacies on soil respiration and microbial community in a Mediterranean forest soil under different soil moisture and carbon inputs[J]. Geoderma, 2022, 405: 115425. doi: 10.1016/j.geoderma.2021.115425 [21] MAIR J, SCHINNER F, MARGESIN R. A feasibility study on the bioremediation of hydrocarbon-contaminated soil from an Alpine former military site: Effects of temperature and biostimulation[J]. Cold Regions Science and Technology, 2013, 96: 122-128. doi: 10.1016/j.coldregions.2013.07.006 [22] GAO Y C, DU J, BAHAR M M, et al. Metagenomics analysis identifies nitrogen metabolic pathway in bioremediation of diesel contaminated soil[J]. Chemosphere, 2021, 271: 129566. doi: 10.1016/j.chemosphere.2021.129566 [23] 刘春柱, 侯萌, 张晴, 等. 长期施入不同量有机肥对农田黑土土壤养分、产量的影响[J]. 中国农学通报, 2017, 33(8): 68-71. LIU C Z, HOU M, ZHANG Q, et al. Effect of long-term application of different amounts of organic fertilizer on black soil nutrients and yield[J]. Chinese Agricultural Science Bulletin, 2017, 33(8): 68-71(in Chinese).
[24] VENTORINO V, PASCALE A, FAGNANO M, et al. Soil tillage and compost amendment promote bioremediation and biofertility of polluted area[J]. Journal of Cleaner Production, 2019, 239: 118087. doi: 10.1016/j.jclepro.2019.118087 [25] WU M L, GUO X Q, WU J L, et al. Effect of compost amendment and bioaugmentation on PAH degradation and microbial community shifting in petroleum-contaminated soil[J]. Chemosphere, 2020, 256: 126998. doi: 10.1016/j.chemosphere.2020.126998 [26] ZHANG D C, MORTELMAIER C, MARGESIN R. Characterization of the bacterial archaeal diversity in hydrocarbon-contaminated soil[J]. Science of the Total Environment, 2012, 421/422: 184-196. doi: 10.1016/j.scitotenv.2012.01.043 [27] MARGESIN R, HÄMMERLE M, TSCHERKO D. Microbial activity and community composition during bioremediation of diesel-oil-contaminated soil: Effects of hydrocarbon concentration, fertilizers, and incubation time[J]. Microbial Ecology, 2007, 53(2): 259-269. doi: 10.1007/s00248-006-9136-7 [28] ATLAS R M, BARTHA R. Microbial ecology: Fundamentals and applications[M]. Pearson Education India, 1998. [29] 顾夏生, 胡洪营, 王慧等. 水处理生物学[M]. 北京: 中国建筑工业出版社, 2010. GU X S, HU H Y, WANG H, et al. Biology for water and wastewater treatment[M]. Beijing: China Architecture & Building Press, 2010(in Chinese).
[30] LAMONACA F, PIZZUTI G, ARCURI N, et al. Monitoring of environmental parameters and pollution by fungal spores in the National Gallery of Cosenza: A case of study[J]. Measurement, 2014, 47: 1001-1007. doi: 10.1016/j.measurement.2013.09.014 [31] 吴蔓莉, 祁燕云, 祝长成, 等. 堆肥对土壤中石油烃的去除及微生物群落的影响[J]. 中国环境科学, 2018, 38(8): 3042-3048. doi: 10.3969/j.issn.1000-6923.2018.08.033 WU M L, QI Y Y, ZHU C C, et al. Influence of compost amendment on hydrocarbon degradation and microbial communities in petroleum contaminated soil[J]. China Environmental Science, 2018, 38(8): 3042-3048(in Chinese). doi: 10.3969/j.issn.1000-6923.2018.08.033
-



 下载:
下载: