-
化学品的生产与使用改善了人类生活质量,但与此同时引发的环境问题和健康危害也不容忽视[1],化学物质可以通过多种途径排放到大气和水体中,母体化合物在环境中发生各种反应,可能会产生毒性更强的污染物[2]. 目前的毒性评价研究更多关注母体化学物质的毒性效应,需进一步研究其在环境中转化产物的毒性和健康效应[3].
吡啶类化合物广泛应用于燃料、农药和医药等生产中[4],被大量排放到环境中,在大气、水和土壤介质中广泛存在. 吡啶是一种高水溶性的含氮芳香族有机化合物,环境中的吡啶类污染物可通过化学和生物方式降解为羟基吡啶[5-6]. 研究表明,许多吡啶化合物具有较强的生物活性,容易诱导急性、亚急性和遗传毒性效应[7]. 遗传毒性特别重要,它与致突变性、致癌性以及癌症有关,氧化应激和DNA损伤修复是遗传毒性中的两条通路,由环境中化学物质引起细胞氧化应激或DNA损伤均可能导致可遗传的性状改变,如细胞突变,可进一步引发癌症[8],有研究表明羟基吡啶可对细胞造成氧化应激,进一步诱导遗传毒性[9]. 突变型和致癌性的生态健康问题越来越凸显,因此需要具有高效率和低成本的遗传毒性筛查评估方法. 大肠杆菌是一种被充分研究的模式生物,基于大肠杆菌的高通量基因毒性评估已经成为DNA损伤修复和氧化应激的重要手段,在环境污染物的暴露下,大肠杆菌中的基因会编码蛋白质进行相关修复,并生成绿色荧光蛋白. 该方法可以检测和量化DNA损伤修复以及氧化应激通路中蛋白质的分子水平变化,涵盖了29种基因标志物,可以对环境污染物进行快速、定量的毒性评估[10-11]. 有机污染物在环境中的降解产物可能表现出比原始污染物更大的毒性效应[12]. 目前,对于吡啶类化合物在光降解过程的中间产物的毒性效应了解甚少,是否会产生毒性更强的转化产物尚未可知,因此运用基于遗传毒性的评价方法评估化学物质的损伤效应能为保护人体健康提供一定的参考价值.
太阳中的紫外线根据波长可以分为UVA(315—400 nm),UVB(280—315 nm)和UVC(200—290 nm),UVC大部分被平流层中的臭氧吸收,到达地面的主要为UVA和UVB[13],2-py、3-py、4-py和2,6-dm-3-py在大气环境中广泛存在,可以在紫外线的作用下发生变化,4种吡啶类化合物对紫外线的吸收峰值距离365 nm和312 nm较近,对这两种波长的紫外线吸收效果较好. 本文利用天然发光菌(费氏弧菌)和重组发光大肠杆菌,研究4种吡啶类化合物经365 nm和312 nm紫外线照射后急性毒性、遗传毒性和氧化应激毒性变化规律.
-
4-羟基吡啶(4-py)、3-羟基吡啶(3-py)、2-羟基吡啶(2-py)、2,6-二甲基-3-羟基吡啶(2,6-dm-3-py)购自阿拉丁公司,具体信息如表1所示. 利用多管涡流器和超纯灭菌水,配制浓度分别为1000、500、200、100、50、20、10、5、2.5、1.25、0.625 mg·L−1C的水溶液.
-
整个光照实验过程在4 oC冰箱中进行. 将装有不同浓度羟基吡啶样品的96孔板放置在距离紫外灯管3 cm处. 分别使用365 nm和312 nm波长的紫外线照射0、1、2、4、8 h,所有样品均保存在4 ℃冰箱内,用于后续毒性测试.
-
费氏弧菌(Vibrio fischeri)在自然状态下会产生光,污染物通过抑制其代谢活动减少荧光强度,因此可通过检测其发光抑制率来评估污染物急性毒性[14]. 本次实验的费氏弧菌冻干粉购自中国海洋文化馆.
将费氏弧菌冻干粉用2% NaCl溶液复苏,在黑色96孔板中每孔加入100 μL菌液,然后再加入100 μL待测样品溶液,设置 2%的NaCl作为空白对照. 将96孔板置于22 °C恒温摇床300 r·min−1孵育30 min,然后使用多功能酶标仪(Synergy Multi-Mode,Biotek)检测费氏弧菌发光强度. 利用公式1计算待测样品和对照组的发光抑制率,每个测试至少设置3个平行样.
EC50是指能引起50%发光抑制效应时对应的暴露浓度,是急性毒性评价中常见的一项指标[15],使用Origin软件(Origin 2021)中的剂量-效应响应曲线拟合吡啶类化合物对费氏弧菌发光抑制的剂量-响应曲线,并计算出50%发光抑制浓度(EC50),即费氏弧菌发光抑制率达到50% 时的样品浓度.
-
本研究所用重组大肠杆菌为K12MG1655[16],每株重组菌携带一个质粒,该质粒含有大肠杆菌功能基因的启动子和绿色荧光蛋白基因(GFP),当重组大肠杆菌暴露于污染物时,其基因表达会受到影响,进而影响启动子和GFP表达,通过检测GFP荧光强度来检测环境污染物对大肠杆菌基因表达的影响[17-18]. 本研究所用重组大肠杆菌来源于Thermo,所选重组大肠杆菌重组基因信息如表2所示.
-
重组E. coli K12 MG1655在37 ℃ LB培养基(1 L溶液中包括10 g胰蛋白胨、5 g酵母提取物和10 g氯化钠)中培养12 h后,将菌液1:100稀释在M9培养基(1 L溶液体系中2 mL 1 mol·L−1 MgSO4、0.1 mL 1 mol·L−1 CaCl2、12.8 g Na2PO4·12H2O、3.0 g KH2PO4、0.5 g NaCl、1.0 g NH4Cl和20 mL 20%葡萄糖)中,以避免LB液体培养基对荧光强度的检测造成干扰. 在M9培养基中培养约2 h后,将处于对数生长期(OD600为0.2—0.3)的重组E. coli转移到透明底的黑色96孔板中,每孔加入180 μL菌液. 同时,迅速将配好的20 μL样品溶液加入重组大肠杆菌菌液中,最终暴露浓度分别为100、20、4、0.8、0.16、0.032、0.0064 mg·L−1C. 空白对照组仅加入M9培养基,每个浓度至少有3个平行样. 在37 ℃培养2 h后,使用多功能酶标仪测量细胞密度OD600(吸光度600 nm)和荧光强度(激发光485 nm,吸收光525 nm)[19].
-
暴露于样品的重组大肠杆菌称为实验组,实验组重组大肠杆菌的荧光强度为Pe,菌体密度表示为ODe,仅加入PBS缓冲液的重组大肠杆菌称为空白对照组,空白对照组大肠杆菌的荧光强度为Pb,菌体密度表示为ODb.
污染物对重组大肠杆菌基因表达的诱导倍数(I),利用公式2计算:
利用转录效应水平指数(TELI)量化和比较不同毒物诱导基因表达之间的差异,TELI利用公式3计算[20]:
其中,Time是指重组大肠杆菌暴露于样品的时间,e是指数学常数e.
-
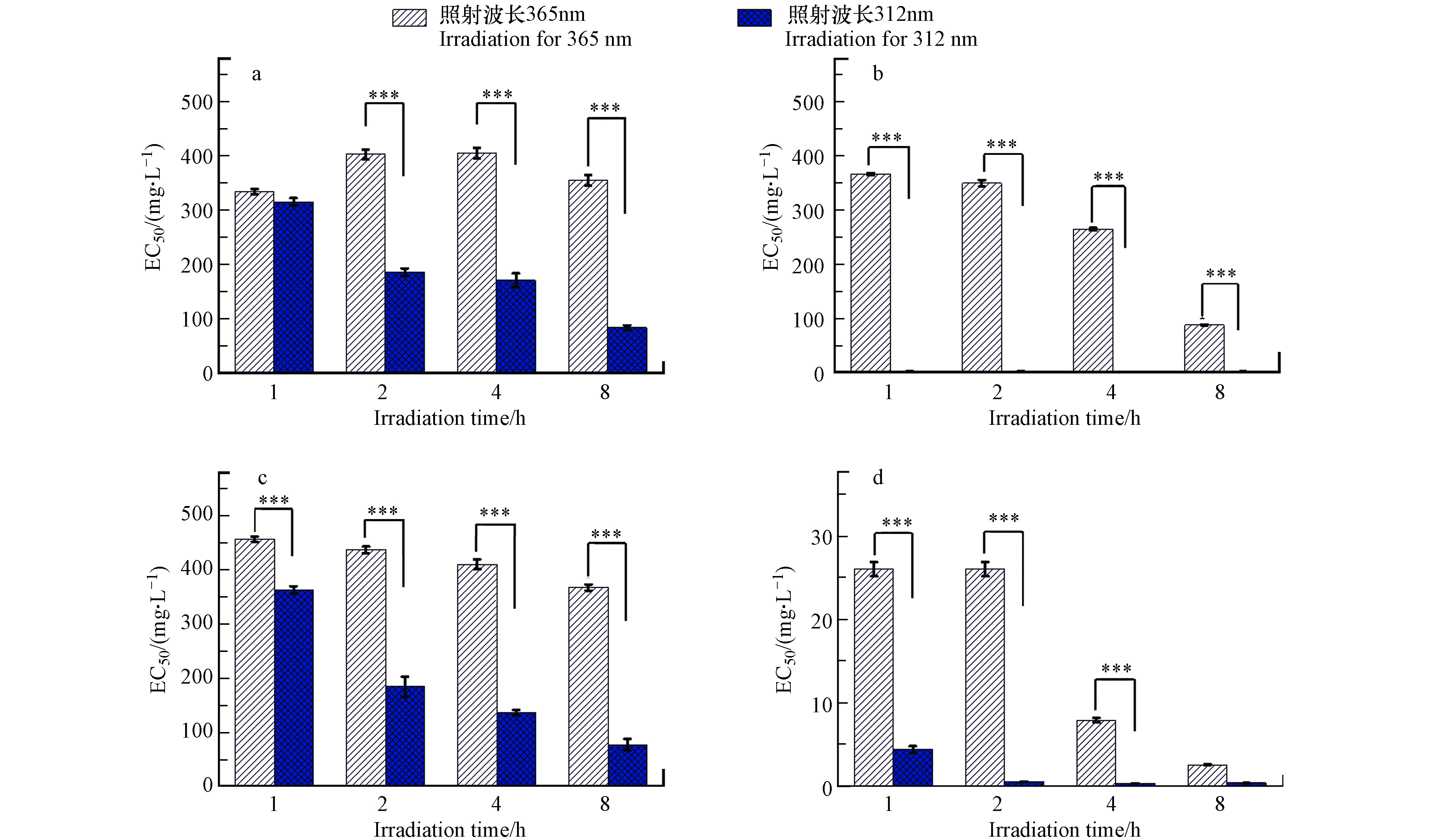
由图1可见,经过365 nm和312 nm 紫外线照射后,4种吡啶化合物的EC50发生了显著变化. 4-py 经365 nm 紫外线照射后EC50小幅升高,而经312 nm 紫外线照射后,4-py的EC50从339.80 mg·L−1C 降低到83.24 mg·L−1C,表明其光降解产物毒性增强,说明4-py经365 nm和312 nm 紫外线照射后产生了不同的降解产物,且312 nm 紫外线照射后降解产物急性毒性显著升高. 有研究表明,草甘膦暴露于365 nm 紫外线后对试验生物的毒性作用没有明显变化,暴露于312 nm 紫外线可造成光解作用降低草甘膦的浓度,从而降低其生态毒性[21].
在所测浓度范围内(64 μg·L−1C—100 mg·L−1C),2-py对费氏弧菌的发光抑制率未达到50%,说明其对费氏弧菌的毒性作用较小,这可能是由于2-py的酸性要弱于其它同分异构体[22],而费氏弧菌一般生活在偏弱碱性的海水中[23],所以其对酸性较小的2-py耐受能力更强. 3-py和2-py经365 nm和312 nm 紫外线照射后,急性毒性均增强. 经312 nm紫外线照射后的毒性增强更为明显,这可能是3-py和2-py经312 nm 紫外线照射后的光降解效率更高造成的. 经365 nm和312 nm 紫外线照射1 h后,2,6-dm-3-py急性毒性急剧增强,这表明2,6-dm-3-py能快速发生光化学反应,且光解产物诱导更强的急性毒性.
-
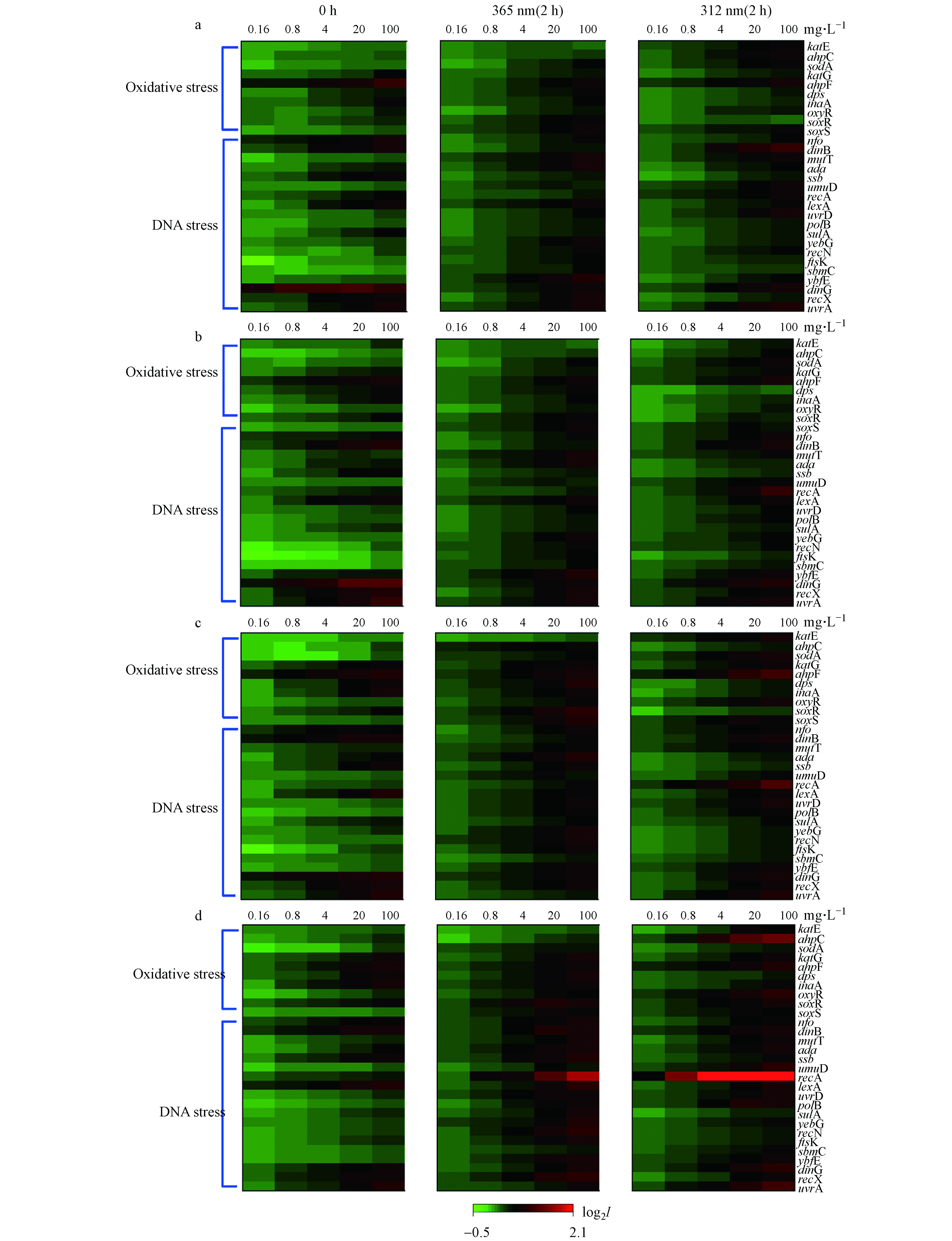
如图2所示,4种吡啶类化合物经紫外线照射后,均可诱导重组大肠杆菌毒性相关基因表达改变,基因表达变化程度会随着浓度增加而增大,有研究报道暴露浓度会影响基因激活程度[24]. 4-py和3-py能够诱导较强的DNA损伤效应,其中lexA、nfo、uvrA基因表达显著增强,但经紫外线照射后,其DNA损伤效应减弱,表明其光降解产物的遗传毒性减弱. 2-py和2,6-dm-3-py对遗传毒性和氧化应激相关基因表达影响较小,但经紫外线照射后,两条通路的相关基因(lexA、recA、oxyR和ahpF等)的表达均被明显激活,表明其光降解产物具有较强的DNA损伤和氧化应激毒性(图2).
4-py具有明显的遗传毒性,可激活lexA基因(参与细菌SOS反应的一种调节蛋白)[25],经紫外线照射后表现出更强的DNA损伤和氧化应激毒性,有一半以上的DNA修复基因(lexA、mutT、uvrA、uvrD和ssb等)和氧化应激相关基因(soxS、soxR和ahpF)显著表达. 3-py经312 nm紫外线照射后,遗传毒性和氧化应激效应更强,可能因为其对312 nm的紫外线吸收能力较强[9].
-
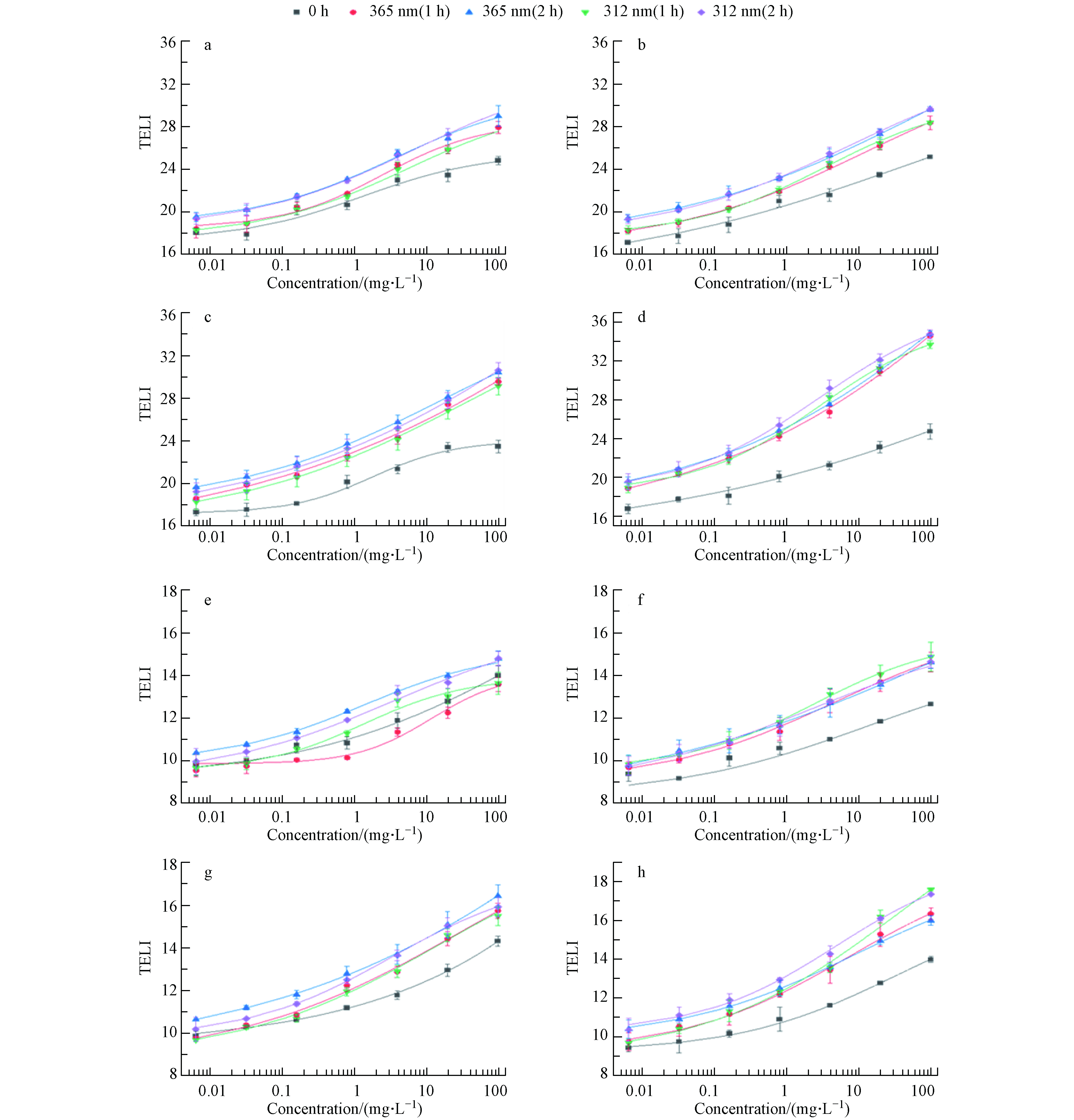
基因表达检测方法从深层次揭示了污染物对生物体遗传信息的影响,探究母体化合物及其降解产物的毒性机制区别. 根据TELI(Transcriptional Effect Level Index)测定的定义和原理,可以计算DNA损伤和氧化应激相关基因的TELI值,以量化各类通路对应的转录水平响应[20].
图3(a、b、c和d)显示了DNA应激相关基因TELI值的剂量响应曲线,这些基因在细胞DNA受到损伤时起到修复的功能. 未经紫外线照射,2-py在DNA应激类别中的TELI最大值为23.4,表明其造成的DNA损伤较弱,其余3种吡啶类化合物TELI值差异不大. 经365 nm 紫外线照射,2-py、4-py和2,6-dm-3-py的TELI值扩大,能激活参与DNA损伤和修复途径的基因(lexA和recA),包括双链断裂修复基因,表明其降解产物能够造成严重的DNA胁迫效应. 4种吡啶类化合物均能激活ada基因,细胞面对DNA损伤时进行直接修复. 2-py、2,6-dm-3-py及其降解产物能够导致参与碱基切除修复基因mutT、nfo、polB表达的上调,表明其降解产物造成了多种结构的DNA损伤. 经312 nm紫外线照射后,3-py的TELI最大值为29.6,是未经照射的1.2倍,导致一些DNA修复通路的基因表达水平发生变化,包括碱基切除修复,核苷酸切除修复等相关基因,表明可能产生了未知基因毒性的降解产物.
RecA和LexA是参与细菌SOS反应的两种重要蛋白,调控DNA修复和突变的基因以响应DNA损伤[25, 26]. mutT、polB、nfo基因参与了碱基切除修复的过程. uvrA能够识别细菌中的损伤,dinG是超家族II解旋酶的成员,参与了核苷酸切除修复和重组DNA修复途径[27],dinB编码特殊的DNA聚合酶应对DNA的断裂[28]. 高度稳定的细菌单链DNA结合蛋白ssb也在定向错配修复中起着关键性的作用,它可以参与双链断裂的损伤反应,促进DNA复制、重组和修复过程[29, 30].
-
图3(e、f、g、h)显示了氧化应激相关基因TELI值的剂量-响应曲线,这些基因在细胞中起到抵制氧化应激的作用,还会通过上调抗氧化及相关酶的合成清除氧化自由基,以消除活性氧对细胞的毒性效应[31]. 未经紫外线照射,3-py在氧化应激类别中的TELI最大值为12.6,表明其引起的氧化应激水平最低,但经312 nm 紫外线照射后,TELI值是未经照射的1.2倍,表明其造成的氧化应激胁迫增强. 2-py未经紫外线照射时表现出显著的氧化应激,这与过往的研究一致[22]. 经紫外线照射后,2-py的TELI最大值由14.3变为16.4,表明其诱导的氧化损伤随光降解作用而增强,可能是由于2-py经紫外线照射后降解产生了杜瓦吡啶酮[23]. 3-py经紫外线照射后TELI值扩大1.3倍,其可作为内源性敏化剂,经紫外线照射对细胞造成光氧化应激,具有细胞抑制作用[25]. 经紫外线照射后,4-py的TELI值变化不大,可能是4-py经紫外线照射后未发生明显降解. 经365 nm 紫外线照射后,2,6-dm-3-py改变了一些氧化应激基因表达,包括oxyR、ahpC、inaA等,表明其降解产物氧化能力增强.
在所选的氧化应激通路相关基因中,oxyR、soxR和soxS是调控活性氧传感器的关键基因,面对外界过氧化氢活性氧的干扰,重组大肠杆菌中oxyR系统会调节相关基因的表达,在感知氧化信号的同时还能上调具有保护细胞免受活性氧干扰的基因产物[32]. ahpC和ahpF是两个参与过氧化物解毒过程的基因,调节烷基氢过氧化物还原酶的生成[33]. katE和katG基因在过氧化氢浓度高的时候起主要作用[34, 35]. sodA基因是保护细胞免受活性氧损害的第一道防线,由它调控的超氧化物歧化酶可以将超氧自由基转化为过氧化氢和水[36]. inaA基因编码一种蛋白酶来应对氧化,但它的诱导依赖于ph[37].
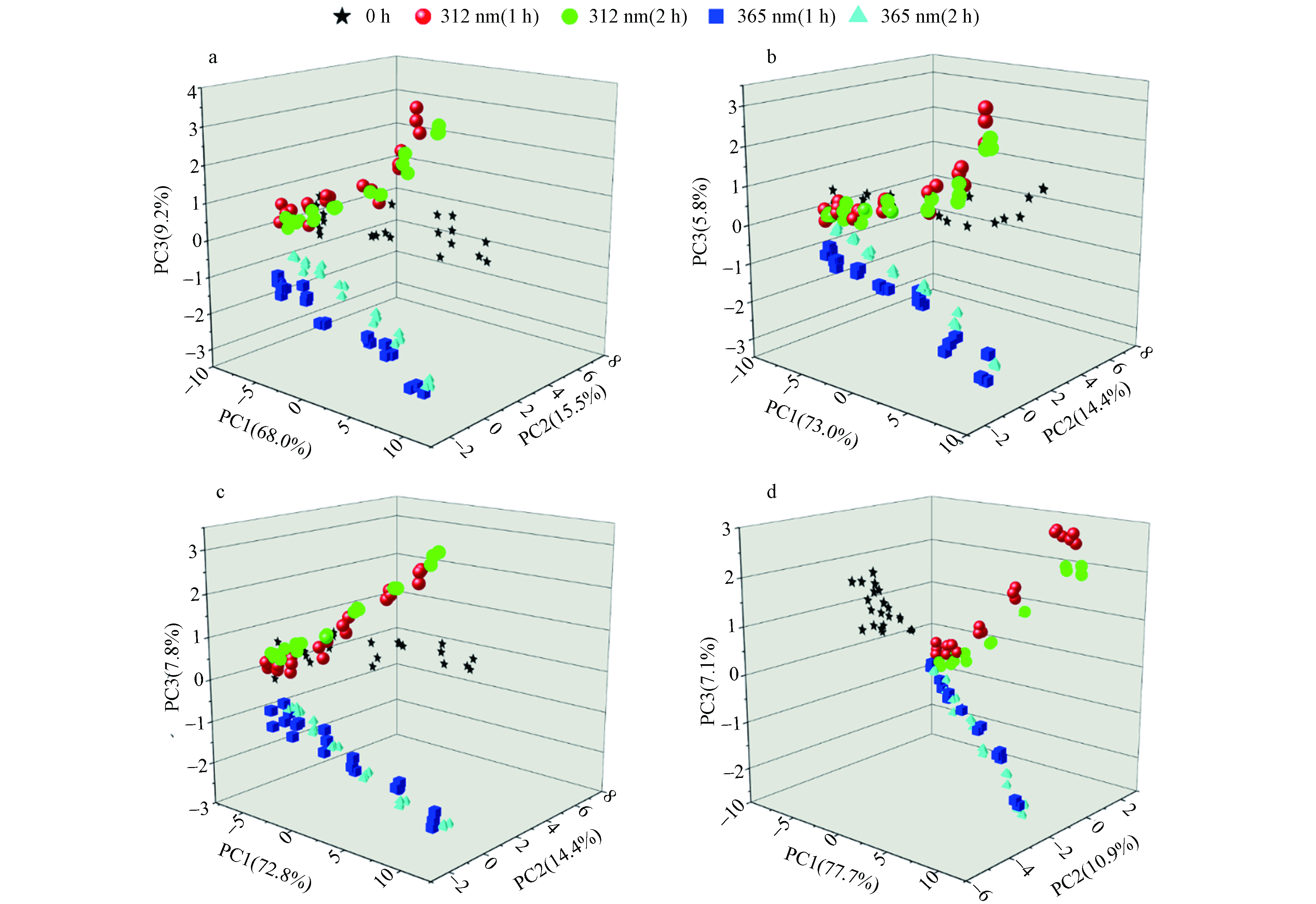
图4为基于4种吡啶类化合物诱导重组大肠杆菌中基因表达数据的PCA分析图. 结果显示,4种吡啶类化合物及其光降解产物的基因表达可以根据遗传毒性机制进行区分. 对于4种吡啶类化合物,紫外线波长、紫外线照射时间和浓度条件都能改变DNA应激和氧化应激相关的基因表达. 如图4所示,未经紫外线照射的点与其他点存在分离,表明紫外线照射能够增强吡啶类化合物诱导DNA损伤的能力. 对于3-py(b),经312 nm紫外线照射1 h和2 h的点距离较近,表明照射时间对其造成的毒性效应影响较小. 对于2,6-dm-3-py,未经紫外线照射的条件下,7个浓度的点聚集在一起,表明其在所选范围浓度内激活相关基因的数量比较相似,造成的损伤效应较弱.
-
(1)羟基吡啶合物光降解产物对费氏弧菌的抑制率随浓度、紫外线波长和辐射时间而变化. 经365 nm和312 nm 紫外线照射后,3-py、2-py和2,6-dm-3-py的EC50随着辐照时间的增加而降低,表明其诱导的毒性作用增强. 经365 nm紫外线照射后,4-py的EC50随着辐照时间先上升后下降. 相反,经312 nm紫外线辐照后,其EC50随辐照时间的增加而降低.
(2)不同的羟基吡啶化合物诱导不同的遗传毒性和氧化应激效应,2-py在DNA应激的TELI值为23.4,只能激活部分DNA损伤修复基因(uvrA和recX等),造成轻微的DNA损伤,3-py在DNA应激的TELI值为25.1,能激活recA、mutT和nfo等基因,造成的DNA损伤较为强烈;2,6-dm-3-py具有显著的遗传毒性和氧化应激效应,能够诱导半数以上的DNA损伤修复和氧化应激毒性通路相关基因(recA、dinG和oxyR等),表明其对重组大肠杆菌具有强烈的胁迫效应.
(3)羟基吡啶化合物经365 nm和312 nm 紫外线照射后,改变DNA应激和氧化应激相关基因表达,其TELI值是未经照射下的1.2—1.4倍,表明其在紫外线辐照作用下的降解产物造成更为严重的DNA损伤和氧化应激毒性.
(4)采用基于DNA损伤和修复途径的遗传毒性评估方法,能够快速、定量筛选以及评估环境中可能对人体造成遗传毒性的化学物质,这种方法更具有成本效应,并且可以获得更多关于DNA损伤和氧化应激潜在机制的信息.
紫外线照射对羟基吡啶毒性效应的影响
Study on mechanism of toxicity of pyridine compounds under ultraviolet irradiation
-
摘要: 吡啶类化合物广泛存在于各种环境介质中,本文以羟基吡啶(2-羟基吡啶(2-py)、3-羟基吡啶(3-py)、4-羟基吡啶(4-py)和2,6-二甲基-3-羟基吡啶(2,6-dm-3-py))为研究对象,利用费氏弧菌和基因重组大肠杆菌,评估4种羟基吡啶经紫外线照射后急性毒性、遗传毒性和氧化应激效应变化规律. 结果表明,2-py、3-py和2,6-dm-3-py经365 nm和312 nm紫外线照射后,毒性随照射时间延长而增强,其急性毒性相较于未照射前,分别增强了24.7%—99.4%和84.0%—99.9%(照射8 h后). 而4-py 经两种波长的紫外线照射后毒性变化不同,经365 nm紫外线照射后,急性毒性降低;但经312 nm紫外线照射8 h后,急性毒性增强77.7%,说明4-py经两种波长紫外线照射后生成了不同的降解产物. 遗传毒性测试结果表明,4种羟基吡啶及其降解产物对DNA损伤和修复过程具有浓度依赖性,在20—100 mg·L−1C浓度下,细胞SOS响应和氧化还原应激相关基因表达显著,但在6.4 —10 mg·L−1C 浓度范围内,毒性相关基因表达变化不大,说明4种羟基吡啶及其降解产物在20—100 mg·L−1 C内可引起DNA损伤和氧化应激效应.Abstract: Pyridine compounds are widely found in various environments. In order to understand the toxic effects in the environment and reveal the influence of ultraviolet (UV) light on their toxicity, the toxicities of 4 pyridine compounds (2-hydroxypyridine (2-py), 3-hydroxypyridine (3-py), 4-hydroxypyridine (4-py) and 2,6-dimethyl-3-hydroxypyridine (2,6-dm-3-py) and their degradation products after UV irradiation were evaluated by both Vibrio fischeri luminescence inhibition assay and genotoxicity assay using recombinant Escherichia coli, respectively. The results showed that the acute toxicities of 2-py, 3-py and 2, 6-dm-3-py increased by 24.7%—99.4% and 84.0%—99.9% after 365 nm and 312 nm UV irradiation for 8 h, respectively. The acute toxicity of 4-py was unchanged after irradiated by 365 nm UV for 8 h, on the contrary, increased by 77.7% after irradiation at 312 nm for 8 h. The DNA damage and repair process induced by the four pyridine compounds and their degradation products was concentration-depended. At the concentration of 20—100 mg·L−1C, the SOS response and Redox stress-related genes were significantly expressed, but at the concentration of 6.4 mg·L−1 to 10 mg·L−1C, the expression of toxicity-related genes showed little change, indicating that the four pyridine compounds and their degradation products at 20—100 mg·L−1C could cause DNA damage and oxidative stress.
-

-
图 2 4种羟基吡啶(a: 4-py, b: 3-py, c: 2-py, d: 2,6-dm-3-py,未经辐照、经365 nm和312 nm紫外线辐照2 h)对重组菌遗传毒性及氧化应激毒性通路相关基因表达谱的影响
Figure 2. Effects of 4 kinds of pyridine compounds (a: 4-py, b: 3-py, c: 2-py, d: 2,6-dm-3-py, unirradiated, irradiated for 2 h with 365 and 312 nm) on the expression of genes involved in genotoxicity and oxidative stress pathways in recombinant bacteria.
图 3 羟基吡啶化合物(a-d: DNA应激相关基因TELI值,其中a: 4-py, b: 3-py, c: 2-py, d: 2,6-dm-3-py,e-f: 氧化应激相关基因TELI值,其中e: 4-py, f: 3-py, g: 2-py, h: 2,6-dm-3-py,未经辐照、经365 nm和312 nm紫外线照射1h、2 h)基于TELI值的剂量响应曲线
Figure 3. Dose response curves(The TELI value of the blank control group during DNA stress was 19, and that of the blank control group during oxidative stress was 10) based on TELI values for 4 kinds of pyridine compounds
表 1 四种羟基吡啶样品
Table 1. Four hydroxypyridine samples
中文名称
Chinese name英文名称
English name简称
Abbreviation分子式
Molecular formulaCAS 4-羟基吡啶 4-pyridone 4-py C5H5NO 626-64-2 3-羟基吡啶 3-pyridone 3-py C5H5NO 109-00-2 2-羟基吡啶 2-pyridone 2-py C5H5NO 142-08-5 2,6-二甲基-3-羟基吡啶 2,6-Dimethyl-3-pyridone 2,6-dm-3-py C7H9NO 1122-43-6 表 2 所选重组大肠杆菌重组基因及其功能
Table 2. Endpoints and functions of genes selected by genotoxicity tests
基因通路
Gene pathways基因
Genes功能
FunctionsDNA应激
DNA stressuvrA 识别细胞内的损伤 recX 调控基因 dinG 超家族II解旋酶之一 ybfE DNA损伤耐受 sbmC 调节基因 ftsK 编码ATP酶 recN 维持染色体结构 yebG 受到lexA基因的调控 sulA 为受损DNA修复提供时间 polB 参与碱基切除修复 uvrD 超家族I DNA解旋酶 lexA 调控DNA修复和突变 DNA应激
DNA stressrecA 调控DNA修复和突变 umuD 参与基因突变 ssb 参与定向错配修复 ada 参与DNA烷基化修复基因转录 mutT 参与碱基切除修复 dinB 编码DNA聚合酶 nfo 参与碱基切除修复 氧化还原应激
Redox stresssoxS 活性氧传感器,编码抗氧化酶 soxR 活性氧传感器,编码抗氧化酶 oxyR 活性氧传感器,保护细胞 inaA 编码蛋白酶 dps 非特异性DNA结合蛋白 ahpF 调节烷基氢过氧化物还原酶 katG 消除过氧化氢 sodA 调控超氧化物歧化酶 ahpC 调节烷基氢过氧化物还原酶 katE 消除过氧化氢 -
[1] LAN J Q, GOU N, RAHMAN S M, et al. A quantitative toxicogenomics assay for high-throughput and mechanistic genotoxicity assessment and screening of environmental pollutants [J]. Environmental Science & Technology, 2016, 50(6): 3202-3214. [2] JONES K C. Persistent organic pollutants (POPs) and related chemicals in the global environment: Some personal reflections [J]. Environmental Science & Technology, 2021, 55(14): 9400-9412. [3] LIU Q F, LI L, ZHANG X M, et al. Uncovering global-scale risks from commercial chemicals in air [J]. Nature, 2021, 600(7889): 456-461. doi: 10.1038/s41586-021-04134-6 [4] ZHANG Y M, CHANG L, YAN N, et al. UV photolysis for accelerating pyridine biodegradation [J]. Environmental Science & Technology, 2014, 48(1): 649-655. [5] SUN J Q, XU L, TANG Y Q, et al. Bacterial pyridine hydroxylation is ubiquitous in environment [J]. Applied Microbiology and Biotechnology, 2014, 98(1): 455-464. doi: 10.1007/s00253-013-4818-9 [6] VASUDEVAN D, DORLEY P J, ZHUANG X. Adsorption of hydroxy pyridines and quinolines at the metal oxide-water interface: Role of tautomeric equilibrium [J]. Environmental Science & Technology, 2001, 35(10): 2006-2013. [7] JO Y W, BIN IM W, RHEE J K, et al. Synthesis and antibacterial activity of oxazolidinones containing pyridine substituted with heteroaromatic ring [J]. Bioorganic & Medicinal Chemistry, 2004, 12(22): 5909-5915. [8] CHU M J, SUN C Q, CHEN W H, et al. Personal exposure to PM2.5, genetic variants and DNA damage: A multi-center population-based study in Chinese [J]. Toxicology Letters, 2015, 235(3): 172-178. doi: 10.1016/j.toxlet.2015.04.007 [9] WONDRAK G T, ROBERTS M J, JACOBSON M K, et al. 3-hydroxypyridine chromophores are endogenous sensitizers of photooxidative stress in human skin cells [J]. Journal of Biological Chemistry, 2004, 279(29): 30009-30020. doi: 10.1074/jbc.M404379200 [10] GOU N, YUAN S H, LAN J Q, et al. A quantitative toxicogenomics assay reveals the evolution and nature of toxicity during the transformation of environmental pollutants [J]. Environmental Science & Technology, 2014, 48(15): 8855-8863. [11] GEURTSEN J, de BEEN M, WEERDENBURG E, et al. Genomics and pathotypes of the many faces of Escherichia coli [J]. FEMS Microbiology Reviews, 2022, 46(6): fuac031. doi: 10.1093/femsre/fuac031 [12] La FARRÉ M, PÉREZ S, KANTIANI L, et al. Fate and toxicity of emerging pollutants, their metabolites and transformation products in the aquatic environment [J]. TrAC Trends in Analytical Chemistry, 2008, 27(11): 991-1007. doi: 10.1016/j.trac.2008.09.010 [13] SOUAK D, BARREAU M, COURTOIS A, et al. Challenging cosmetic innovation: The skin microbiota and probiotics protect the skin from UV-induced damage [J]. Microorganisms, 2021, 9(5): 936. doi: 10.3390/microorganisms9050936 [14] PIPES B L, NISHIGUCHI M K. Nocturnal acidification: A coordinating cue in the Euprymna scolopes- Vibrio fischeri symbiosis [J]. International Journal of Molecular Sciences, 2022, 23(7): 3743. doi: 10.3390/ijms23073743 [15] STEPNIEWSKA K, CHOTIVANICH K, BROCKMAN A, et al. Overestimating resistance in field testing of malaria parasites: Simple methods for estimating high EC50 values using a Bayesian approach [J]. Malaria Journal, 2007, 6: 4. doi: 10.1186/1475-2875-6-4 [16] AGARWAL A, ALLAMANENI S S R. Sperm DNA damage assessment: A test whose time has come [J]. Fertility and Sterility, 2005, 84(4): 850-853. doi: 10.1016/j.fertnstert.2005.03.080 [17] JANG J, HUR H G, SADOWSKY M J, et al. Environmental Escherichia coli: Ecology and public health implications-a review [J]. Journal of Applied Microbiology, 2017, 123(3): 570-581. doi: 10.1111/jam.13468 [18] MOREB E A, HOOVER B, YASEEN A, et al. Managing the SOS response for enhanced CRISPR-cas-based recombineering in E. coli through transient inhibition of host RecA activity [J]. ACS Synthetic Biology, 2017, 6(12): 2209-2218. doi: 10.1021/acssynbio.7b00174 [19] REIFFERSCHEID G, BUCHINGER S. Cell-based genotoxicity testing: Genetically modified and genetically engineered bacteria in environmental genotoxicology [J]. Advances in Biochemical Engineering/Biotechnology, 2010, 118: 85-111. [20] GOU N, GU A Z. A new Transcriptional Effect Level Index (TELI) for toxicogenomics-based toxicity assessment [J]. Environmental Science & Technology, 2011, 45(12): 5410-5417. [21] PAPAGIANNAKI D, MEDANA C, BINETTI R, et al. Effect of UV-A, UV-B and UV-C irradiation of glyphosate on photolysis and mitigation of aquatic toxicity [J]. Scientific Reports, 2020, 10: 20247. doi: 10.1038/s41598-020-76241-9 [22] SKOUTELIS C G, VLASTOS D, KORTSINIDOU M C, et al. Induction of micronuclei by 2-hydroxypyridine in water and elimination of solution genotoxicity by UVC (254 nm) photolysis [J]. Journal of Hazardous Materials, 2011, 197: 137-143. doi: 10.1016/j.jhazmat.2011.09.065 [23] STAPLETON D R, KONSTANTINOU I K, KARAKITSOU A, et al. 2-Hydroxypyridine photolytic degradation by 254 nm UV irradiation at different conditions [J]. Chemosphere, 2009, 77(8): 1099-1105. doi: 10.1016/j.chemosphere.2009.08.026 [24] NORTH M, TANDON V J, THOMAS R, et al. Genome-wide functional profiling reveals genes required for tolerance to benzene metabolites in yeast [J]. PLoS One, 2011, 6(8): e24205. doi: 10.1371/journal.pone.0024205 [25] MO C Y, CULYBA M J, SELWOOD T, et al. Inhibitors of LexA autoproteolysis and the bacterial SOS response discovered by an academic-industry partnership [J]. ACS Infectious Diseases, 2018, 4(3): 349-359. doi: 10.1021/acsinfecdis.7b00122 [26] SCHLACHER K, PHAM P, COX M M, et al. Roles of DNA polymerase V and RecA protein in SOS damage-induced mutation [J]. Chemical Reviews, 2006, 106(2): 406-419. doi: 10.1021/cr0404951 [27] REN B B, DUAN X W, DING H G. Redox control of the DNA damage-inducible protein DinG helicase activity via its iron-sulfur cluster [J]. Journal of Biological Chemistry, 2009, 284(8): 4829-4835. doi: 10.1074/jbc.M807943200 [28] UCHIDA K, FURUKOHRI A, SHINOZAKI Y, et al. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal [J]. Molecular Microbiology, 2008, 70(3): 608-622. doi: 10.1111/j.1365-2958.2008.06423.x [29] CUBEDDU L, WHITE M F. DNA damage detection by an archaeal single-stranded DNA-binding protein [J]. Journal of Molecular Biology, 2005, 353(3): 507-516. doi: 10.1016/j.jmb.2005.08.050 [30] LU D, WINDSOR M A, GELLMAN S H, et al. Peptide inhibitors identify roles for SSB C-terminal residues in SSB/exonuclease I complex formation [J]. Biochemistry, 2009, 48(29): 6764-6771. doi: 10.1021/bi900361r [31] KESELER I M, COLLADO-VIDES J, SANTOS-ZAVALETA A, et al. EcoCyc: A comprehensive database of Escherichia coli biology [J]. Nucleic Acids Research, 2011, 39(Suppl_1): D583-D590. [32] CHRISTMAN M F, STORZ G, AMES B N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins [J]. Proceedings of the National Academy of Sciences of the United States of America, 1989, 86(10): 3484-3488. doi: 10.1073/pnas.86.10.3484 [33] OCHSNER U A, VASIL M L, ALSABBAGH E, et al. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF [J]. Journal of Bacteriology, 2000, 182(16): 4533-4544. doi: 10.1128/JB.182.16.4533-4544.2000 [34] KAKU N, HIBINO T, TANAKA Y, et al. Effects of overexpression of Escherichia coli katE and bet genes on the tolerance for salt stress in a freshwater Cyanobacterium Synechococcus sp. PCC 7942 [J]. Plant Science, 2000, 159(2): 281-288. doi: 10.1016/S0168-9452(00)00353-8 [35] KARIMOV I F, DERYABIN D G, KARIMOVA D N, et al. Evaluation of oxidative metabolism in leukocytes during phagocytosis of Escherichia coli carrying genetic constructs soxS: Lux or katG: Lux [J]. Bulletin of Experimental Biology and Medicine, 2016, 161(2): 276-280. doi: 10.1007/s10517-016-3394-2 [36] ZHANG M M, QIN Y X, HUANG L X, et al. The role of sodA and sodB in Aeromonas hydrophila resisting oxidative damage to survive in fish macrophages and escape for further infection [J]. Fish & Shellfish Immunology, 2019, 88: 489-495. [37] LEE S, MITCHELL R J. Detection of toxic lignin hydrolysate-related compounds using an inaA: LuxCDABE fusion strain [J]. Journal of Biotechnology, 2012, 157(4): 598-604. doi: 10.1016/j.jbiotec.2011.06.018 -



 下载:
下载: