-
多环芳烃(PAHs)是一类由2个或2个以上苯环在线性、角形或簇状排列下组成的半挥发性有机化合物[1],包括萘(NAP)、蒽(ANT)、菲(PHE)、芘(PYR)和苯并[a]芘(BaP)等在内的150余种。PAHs的低生物利用率、疏水性和热力学稳定性等特征,使其在环境中的降解率较低[2]。
PYR作为典型的四环PAHs,常被用作高环芳烃降解的研究基础。细菌可以在有氧或无氧条件下代谢PYR。PYR的有氧代谢已得到广泛研究,尤其是在分枝杆菌和解环菌中较为常见[3]。在有氧降解途径中,PYR在单加氧酶或双加氧酶的催化下被氧化。双加氧酶是一种复杂的多组分酶体系,由铁氧还蛋白、还原酶和含铁硫的末端加氧酶组成[4]。PYR的苯环可以被双加氧酶羟基化形成顺式二氢二醇,然后被脱氢酶氧化为次二醇,这些次二醇被内环裂解双加氧酶通过邻位裂解,或通过外环裂解双加氧酶进行间位裂解,生成常见的邻苯二酚,然后转化为三羧酸循环(TCA)所需要的代谢物质[3,5],这是最常见的PYR细菌降解途径。其中涉及到的酶主要包括双加氧酶、二氢庚二醇脱氢酶、断裂双加氧酶、乙醇脱氢酶、乙醛脱氢酶和脱羧酶等。
目前发现能够降解PYR的典型细菌有假交替单胞菌属(PseudoAlteromonas sp.)[6]、解环菌属(Cyclocasticus sp.)[7-8]、芽孢杆菌属(Bacillus sp.)[9-12]、红球菌属(Rhodococcus sp.)[13]、黄杆菌属(Flavobacterium sp.)[14-15]等。本文利用PYR作为唯一碳源,在被石油烃污染码头的海底沉积物中分离到一株海洋PYR降解菌株,对其进行了分类鉴定,并对降解途径和基因组进行了初步分析,以丰富芳烃化合物海洋降解菌库,以此为海洋环境中PAHs污染治理等提供数据参考。
-
主要试剂: PYR(≥98%)购自美国AccuStandard公司。无机盐培养基[16]包括19 g·L−1NaCl,7.0 g·L−1 MgSO4·7H2O,3.0 g·L−1Na2HPO4,2.0 g·L−1KH2PO4,1.0 g·L−1NH4NO3,0.7 g·L−1KCl 以及微量元素(0.06 g·L−1 MnSO4·H2O,0.5 g·L−1 FeSO4·7H2O和0.2 g·L−1 ZnSO4·7H2O)。富集液体培养基:无机盐培养基中加入5 g·L−1蛋白胨,1 g·L−1酵母粉。富集固体培养基和无机盐固体培养基分别为富集液体培养基和无机盐培养基中添加2%的琼脂。
上述培养基均将pH调节为7.0,在121 ℃下高压蒸汽灭菌20 min后使用。降解培养基和筛选固体培养基为分别在无机盐培养基和无机盐固体培养基中加入PYR-正己烷溶液,并挥发除去正己烷后制成。
-
取5 mL沉积物悬浊液加入45 mL降解培养基中,在避光、30 ℃、150 r·min−1条件下振荡培养,培养7d后,取出5 mL接种于新的更高浓度的PYR驯化培养基中,如此反复共驯化5代,PYR的质量浓度梯度为1、5、10、20、40 mg·L−1,驯化结束之后,在筛选固体培养基上涂布100 μL上述菌液,放置于培养箱中于30 ℃避光培养。3~7 d长出明显菌落后,用划线分离法纯化出单菌株。菌种-80 ℃下用20%甘油保存。
-
选取1株生长最好的菌株命名为P7,将菌株P7单菌落在筛选固体培养基上划线,30 ℃恒温培养,7 d后进行观察并收集菌体进行SEM观测。利用基因组提取试剂盒提取菌株的总DNA,以其为模板,用27F和1492R对DNA进行扩增。PCR反应体系:模板2 μL,KOD OneTM PCR Master Mix 25 μL,1×引物(10 μM each)1.5 μL,灭菌水20 μL。PCR循环条件:变性98 ℃,10 s,退55 ℃,8 s,延伸68 ℃,2 s,经历35个循环后,通过1%琼脂糖凝胶电泳验证PCR产物。产物由科学指南针公司进行测序,测序结果在NCBI网站(http:∥www.ncbi.nlm.nih.gov)上进行比对,若菌株在模式菌株数据库中比对相似度略低,则利用nt数据库进行1次比对。利用菌株P7的16S rDNA的通用引物进行扩增,得到1 361bp的基因序列,测序结果已提交GenBank,登录号为ON573337。
-
首先将P7菌株单菌落接种到富集液体培养基中,30 ℃,150 r·min−1富集到对数生长期(OD600≈1),取30 mL培养物,5000 r·min−1低速离心收集菌体,并用灭菌无机盐培养基洗涤2次,重悬后接入含有PYR的18 mL无机盐培养基中(PYR的终质量浓度为20 mg·L−1,同时设置接种失活菌的培养基作为空白对照)。将上述实验组和空白对照组于30 ℃,150 r·min−1,避光培养。
每24 h取培养液测定OD600,每2 d测定培养液中剩余PYR浓度。用20 mL正己烷分2次萃取培养物中残余的PYR,取1 mL萃取液经0.22 μm滤膜过滤后进行GC-MS分析,最终根据实验组和对照组培养基中PYR的残留量进行降解率的计算,降解率=(对照组PYR浓度-实验组PYR浓度)/对照组PYR浓度。
GC/MS分析条件如下: 色谱条件:Agilent 19091S-433 HP-5MS色谱柱(30 m×250 μm×0.25 μm);进样口250 ℃,检测器280 ℃;初始温度40 ℃,以8 ℃·min−1升温至300 ℃并保持10 min;载气(高纯氦气)流速1.0 mL·min−1,压力7.01 psi;进样量1 μL,分流比20:1。质谱条件:离子源为电子轰击源,电离电压70 eV,温度230 ℃;采集方式为选择离子;溶剂延迟5 min。
-
无机盐培养基中加入一定量的PYR-正己烷溶液,在接种菌液之后调节PYR质量浓度分别为1、5、10、20和40 mg·L−1。接种OD600≈1的P7菌悬液,30 °C、150 r·min−1条件下振荡培养,以测定底物浓度对于菌株P7降解率的影响。PYR质量浓度为20 mg·L−1的降解培养基,接种1%的菌悬液后置于温度为4、15、20、30和35 ℃条件下振荡(150 r·min−1)培养,用于测定温度对于菌株P7降解率的影响。用缓冲体系调节PYR质量浓度为20 mg·L−1的降解培养基的pH分别为4、6、7、8和10,接种1%的菌悬液,30 °C下振荡(150 r·min−1)培养,用于测定pH对于菌株P7降解率的影响。在PYR质量浓度为20 mg·L−1的降解培养基中,加入一定量的NaCl,分别配制成盐度为10%、20%、30%、40%的液体培养基。接种1%的菌悬液,30 °C下振荡(150 r·min−1)培养,用于测定盐度对于菌株P7降解率的影响。20 d后整瓶萃取测定剩余PYR浓度,实验设灭活菌作为对照,每个处理设3个重复。
-
选取P7降解后的样品进行代谢产物的分析测定。采用等体积的正己烷对菌液萃取2次,将萃取液合并后经无水硫酸钠除去水分,取1 mL经0.22 μm滤膜过滤后,利用GC-MS检测。初始温度 40 ℃,以10 ℃·min−1升至240 ℃并保持2 min,后以5 ℃·min−1升至260 ℃并保持1 min,再以3 ℃·min−1升至300 ℃。质谱的采集方式为全扫描。
-
将菌株P7接入PYR降解培养基中,于30 ℃振荡培养10 d后离心,收集沉淀菌体。用磷酸盐缓冲液将菌体重悬后,在冰浴中超声破碎,4 ℃下冷冻离心,离心速度为10 000 r·min−1,取上清液作为粗酶液。以接种前的菌株的粗酶液作为对照。蛋白含量用Bradford法测定。
采用紫外分光光度法,测定菌株的酶活力。测定体系为0.2 mL粗酶液、2.4 mL磷酸缓冲液、0.4 mL 20 μmol·L−1邻苯二酚溶液,混合均匀后,37 ℃水浴加热反应。邻苯二酚2,3-双加氧酶酶活测定:反应30 min后,测定反应液在375 nm波长光吸收的增加值。邻苯二酚2,3-双加氧酶活单位:在30 ℃时,每分钟生成1 μmol·L−12-羟基粘糠半醛所需的酶量[17]。
-
对收集好的样品进行DNA的提取,并对抽提的样品送往科学指南针公司进行检测。采用试剂盒说明书中的标准Illumina TruSeq Nano DNA LT文库制备实验流程构建所需的基因组上机文库。本研究将拼接的而得的序列与数据库NR、COG和KEGG进行比对,得到与给定序列具有最高序列相似性的核酸或蛋白质,获得对应的注释信息。
-
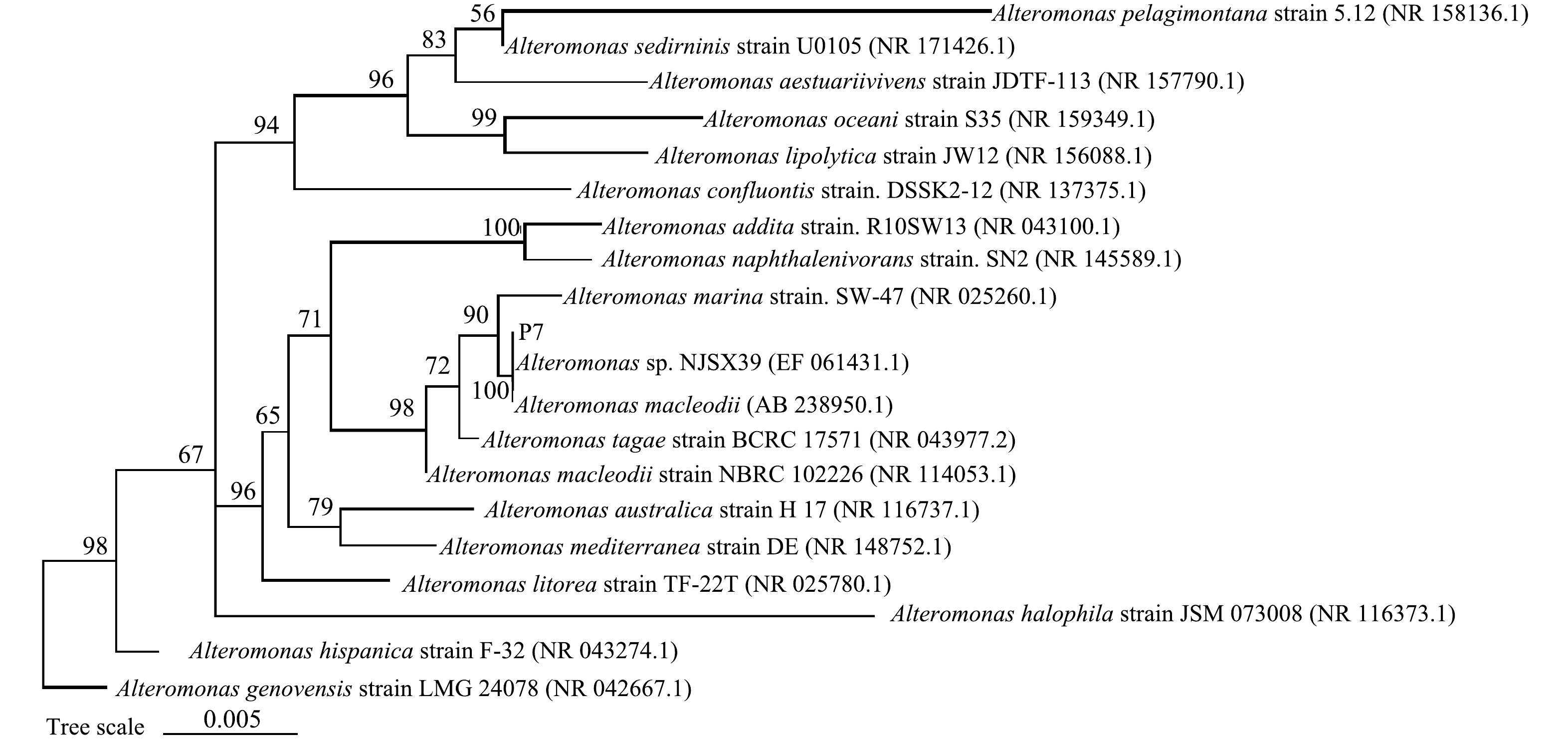
在青岛小港码头海底沉积物中筛选到一株高效PYR降解菌,命名为P7,P7在富集固体培养基上的菌落大小为1~1.5 mm,菌落形态为规则圆形,不透明并呈现白色,表面湿润,边缘整齐,只在培养基表面生长,边缘规则,不向四周扩散。SEM结果如图1所示,菌株P7呈杆状,长度为10~20 μm,无鞭毛。通过相似性比较,根据发育树(图2)并结合各个菌株的菌落形态以及生理生化特征可以得出,菌株P7为交替单胞菌(Alteromonas)。
程晓宇[18]等在东海沉积物中也筛选到了2株未被报道过的具有香草醛降解功能的交替单胞菌,此类种属细菌在海洋环境中广泛分布,并且绝大部分具有潜在的烃类化合物降解能力。但未曾有过关于交替单胞菌降解HMW-PAHs的报道,本文首次筛选分离到了具有HMW-PAHs降解能力的海洋交替单胞菌。目前对海洋中HMW-PAHs的降解菌的筛选研究报道很少。对于Alteromonas sp. P7的分离鉴定可以进一步丰富芳烃化合物海洋降解菌库,并可为原位高效修复海洋PAHs污染提供菌种储备。
-
图3展示了菌株P7的生长曲线和对PYR的降解效果。P7菌株在接种入含有PYR的无机盐培养基之后OD600相对稳定,菌株在此期间逐渐适应营养物质的改变;在第4 天开始快速增长,OD600增加;到第6天时,细菌的OD600值达到最大,为0.163,且在5~8 d内基本稳定,9~20 d内OD600下降,此时培养基中的底物不足以支撑菌株继续增殖,并且存在死亡现象,培养基中出现的絮状物可能是死亡菌体与细菌分泌物的混合体。PYR在前4 d被消耗地比较缓慢,在4~12 d PYR被快速降解,在12d时PYR的质量浓度下降到10.34 mg·L−1,降解率为42.5%,之后PYR的浓度继续降低,但是降解速度逐渐变慢,培养到 20 d 时,PYR的降解率为44.8%。
CAO等[19]从中国渤海石油污染的沉积物中分离出来的pusillimonas sp. T7-7,对PYR的降解率达到90%。从沙特阿拉伯海湾分离到的AchroMobacter Xylosoxidans Py4-B,对PYR的降解率达到80%[20]。Novosphingobium是一种α-变形菌属,首次从韩国蔚山湾分离出来,N. pentaromativorans可以在8 d内降解代谢88.2%~99.9%的PYR[21];LYU等[22]从蔚山湾的沉积物中分离出属于该属的另一株N. pentaromativorans US6-1,其在36 h内对PYR的降解率为31.81% 。与典型的海洋PYR降解菌相比,Alteromonas sp. P7对PYR的降解率相对较低。这可能因为菌株在降解PYR过程中产生了有毒物质,从而抑制细菌的生长以及进一步降解,菌株在降解终点时的OD600值小于初始值也可支持该结论。
-
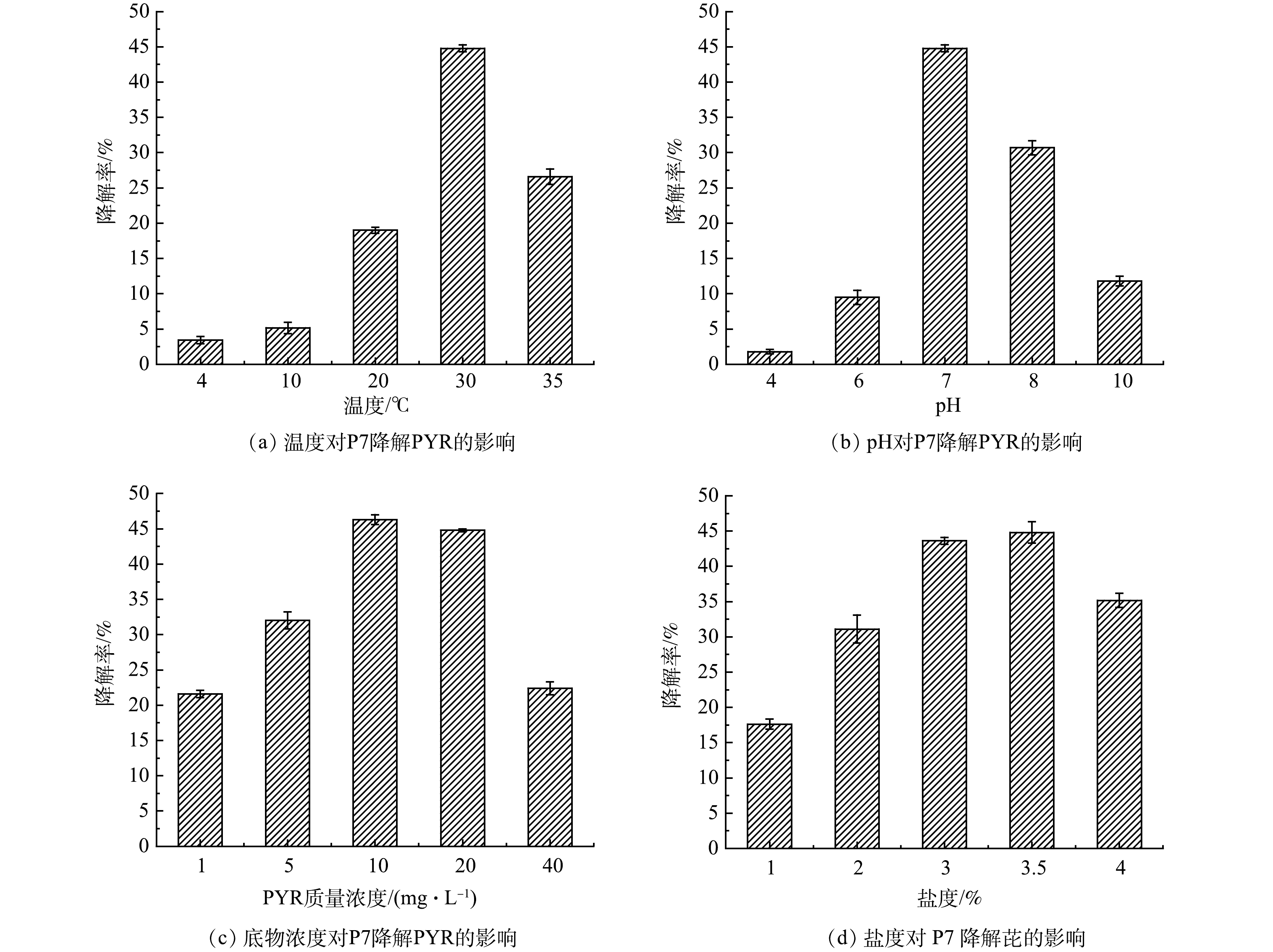
温度不仅能够影响PYR在水中的溶解度,还能够影响细菌对PYR降解的相关酶系的活性,进而影响PYR的降解性能。如图4(a)所示,随着温度升高,菌株P7对PYR的降解率逐渐升高,且在30 ℃降解效果最好,降解率为44.8%。当温度继续上升至35 ℃,降解率略有下降,在4 ℃和10 ℃的条件下,由于酶活性大幅度降低,导致菌株的降解能力变差。
图4(b)反映了PYR初始浓度对于P7降解率的影响。在1~10 mg·L−1内,随着底物浓度的增加,菌株P7对于PYR的降解率增加,当PYR的初始质量浓度分别为1、5和10 mg·L−1时,P7的降解率分别为21.6%、32%、46.3%;随着PYR浓度的升高,降解率有所下降,PYR的初始质量浓度为40 mg·L−1时,菌株P7降解率下降为22.4%。菌株P7在PYR质量浓度为10 mg·L−1时降解率达到最大,为46.3%。PYR浓度过低时,菌株缺少可利用的碳源,导致生长增殖缓慢,降解效果不佳。而PYR浓度过高时,细菌代谢过程中可能会产生毒性物质,从而抑制其降解,且部分PYR不溶于水难以被细菌所利用。
由图4(c)可以看出,在不同pH下,菌株P7对PYR的降解率差异较大。在中性和弱碱性条件下,降解率相对较高,在pH为7.0和8.0时,菌株P7的降解率分别为44.8%和30.7%;酸性条件下,降解率较低,pH为4.0和6.0时,降解率只有1.8%和9.5%;在碱性条件下,菌株P7仍对PYR具有降解效果,在pH为10.0时PYR降解率为11.8%。
盐度在细菌生长的过程中,对调节渗透压平衡起到重要作用,从而影响对PYR的降解能力[23]。如图4(d)所示,菌株P7耐盐度范围广泛,在10%~40%的盐度下,对PYR均有一定的降解作用;在盐度为标准海水盐度(35%)时,菌株P7对PYR的降解率最高,为44.8%。菌株P7的酶活条件和渗透压等,使其在高盐度下仍具有较高的代谢活性。
-
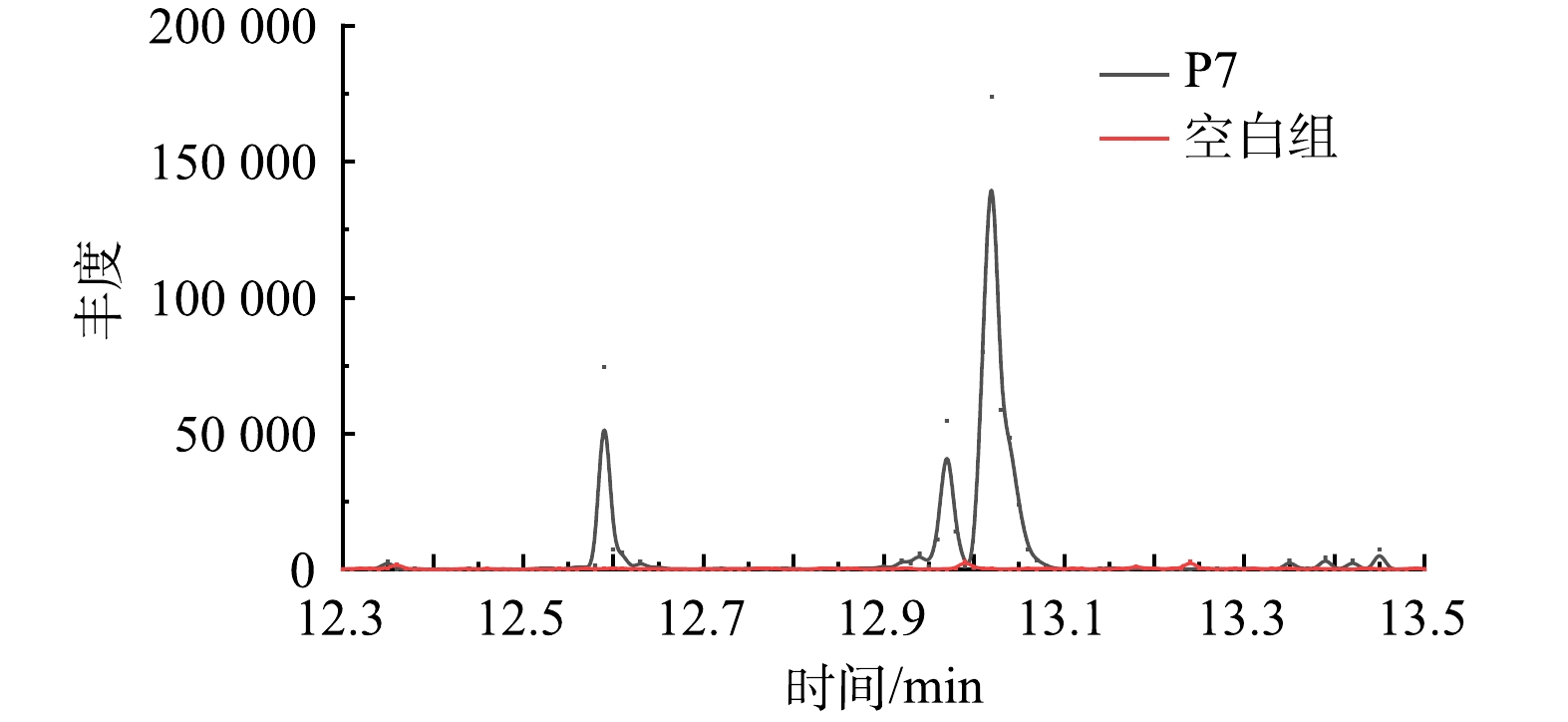
通过GC-MS鉴定了Alteromonas sp. P7降解PYR过程中产生的中间代谢产物。由图5可见,与空白对照对比,Alteromonas sp. P7的反应体系中在第8天后的样品中发现了3种物质,保留时间在12.532、12.928和14.039 min的吸收峰经质谱对比分别为水杨酸、邻苯二甲酸和PYR。
水杨酸和邻苯二甲酸的存在说明Alteromonas sp. P7对PYR的降解存在2条下游途径,分别为邻苯二甲酸途径及水杨酸途径。多条降解途径有利于增强降解菌对于PYR的降解效果,并可以抵制环境的不良影响。
-
PAHs的种类繁多,不同种类微生物,对于PAHs的降解途径可能存在差异,因此,涉及的酶种类众多[24-27]。其中,较为常见的有邻苯二酚双加氧酶,该加氧酶以氧作为底物,可以催化裂解邻苯二酚(C6H4(OH)2)的芳环[28]。邻苯二酚双加氧酶根据芳香环的裂解位置分为2类:邻苯二酚1,2-双加氧酶和邻苯二酚2,3-双加氧酶。邻苯二酚1,2-双加氧酶催化2个羟基之间的碳-碳键的氧化,称为内二醇裂解,产生顺式、顺式-粘康酸。邻苯二酚2,3-双加氧酶催化2个羟基外的碳-碳键的氧化,称为外二醇裂解,产生2-羟基粘醛[29-30]。
结果表明,Alteromonas sp. P7具有邻苯二酚2,3-双加氧酶活力,P7在富集培养基增殖后,邻苯二酚2,3-双加氧酶活力为0.231 μmol·(min·mg)−1,经芘诱导后,邻苯二酚2,3-双加氧酶活力增加到0.537 μmol·(min·mg)−1。由此可以推测菌株P7将代谢产物中的水杨酸或邻苯二甲酸氧化为邻苯二酚后,外断裂邻苯二酚2,3-双加氧酶对邻苯二酚2个羟基之旁开环(间位途径),形成丙酮酸和乙醛,最终进入三羧酸循环,从而实现对PYR的最终降解。
-
采用A5-MiSeq[31]和SPAdes[32]对去除接头序列的测序数据进行从头拼装,构建contig和scaffold。拼装好的Alteromonas sp. P7全基因组总序列长度为4 597 467 bp,GC含量百分比为44.26%。采用GeneMarkS软件对细菌基因组的蛋白质编码基因进行预测,P7开放阅读框数量为4 079,开放阅读框所占碱基数量为4 003 854 bp,基因编码密度为0.887基因·kb−1。对P7非编码RNA进行预测,对于细菌,非编码RNA(ncRNA)的类型主要为sRNA、tRNA和rRNA,其中tRNA含量最高,占基因组比例为0.1007。非编码RNA预测的结果见表1。
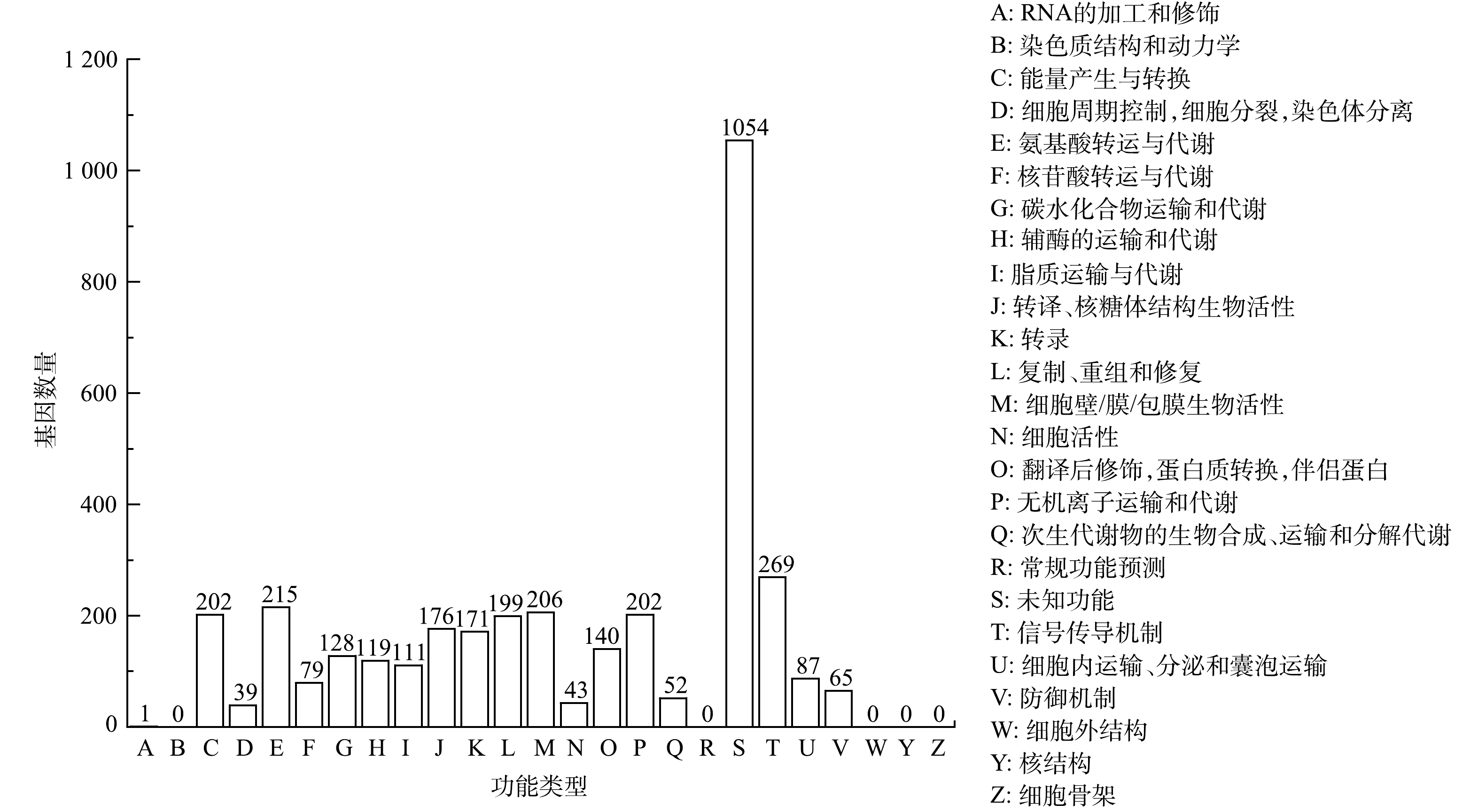
1)蛋白编码基因的COG注释分析。eggNOG(COG)是evolutionary genealogy of genes: non-supervised orthologous groups的缩写[33]。构成每个eggNOG的蛋白都是被假定为来自于一个祖先蛋白,并且是直系同源(orthologs)或者是旁系同源(paralogs)。由图6可知,菌株Alteromonas sp. P7基因组共注释到3 556个编码基因,占总编码基因的87.22%;其中包含大量未知功能的基因,说明可以进一步探索和挖掘这些基因的功能。已知编码功能的基因中,与信号转导机制功能相关基因最多,有269个,占编码基因总数的6.59%;比较多的富集基因类群还包括能量产生与转换、氨基酸运输与代谢、复制、重组和修复、细胞壁/膜/包膜生物活性、无机离子运输有关的基因较多。这可能与Alteromonas sp. P7以PYR为唯一能源进行能量转换有关。
2)蛋白编码基因的KEGG注释分析。KEGG(kyoto encyclopedia of genes and genomes),又称京都基因与基因组百科全书,是基因组破译方面的数据库。对菌株Alteromonas sp. P7在KEGG数据库共注释到的基因共有2 093个,占总编码基因的51.31%。
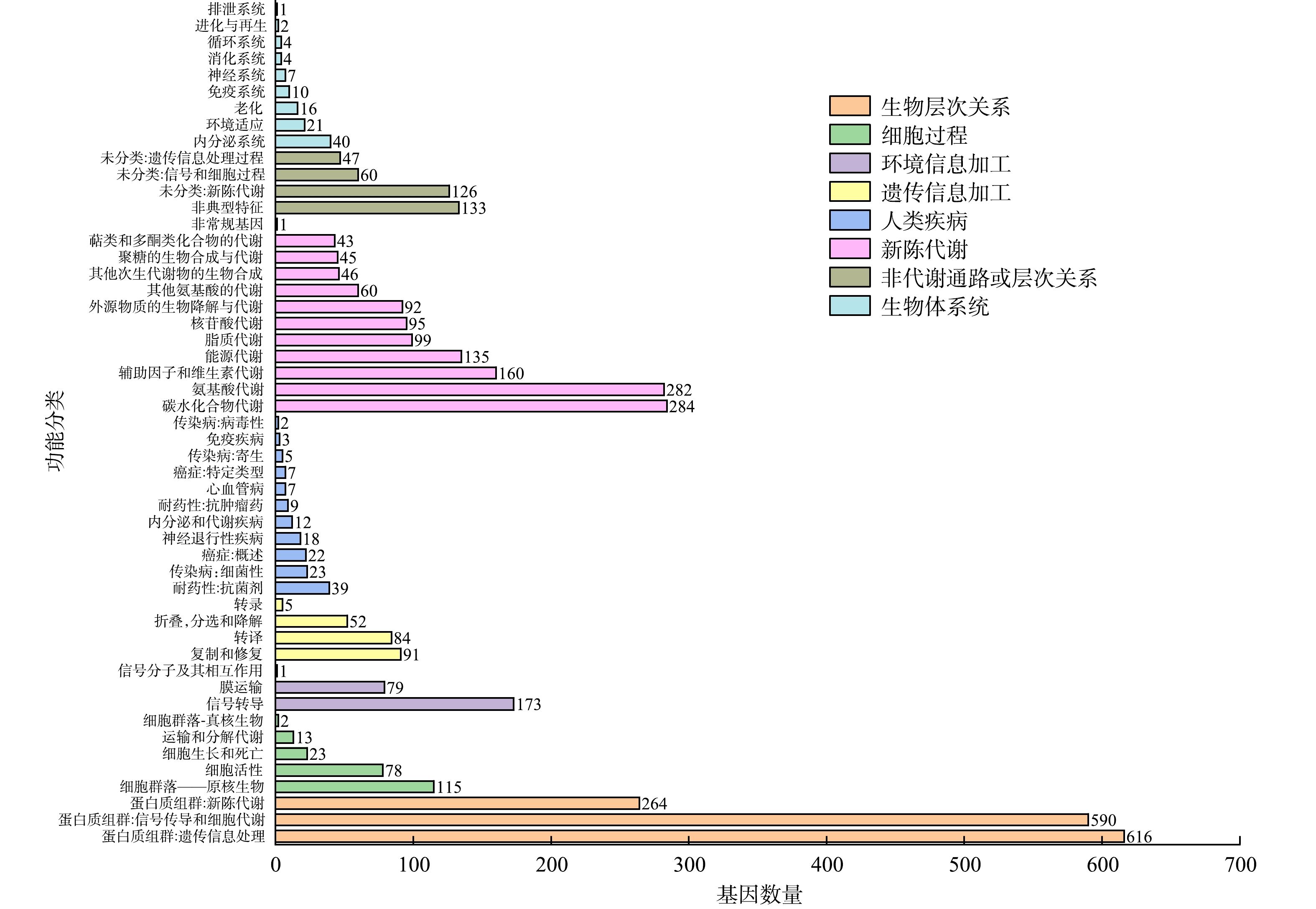
图7为菌株P7的KEGG谢通路图。对Alteromonas sp. P7的蛋白编码基因的KEGG注释分析,发现其中包含最多的有与遗传信息处理相关蛋白质组群和与信号传导和细胞代谢有关的蛋白质组群,分别有616个和590个;涉及的代谢通路有1 342个,其中较多的有碳水化合物代谢、氨基酸代谢、辅助因子和维生素代谢以及能源代谢等类群的相关基因,分别为284、282、160和135个。Alteromonas sp. P7具有21个参与环境适应的基因,将这部分基因与参与碳水化合物代谢的基因和参与外源生物降解与代谢过程的基因联系起来研究,可以进一步为阐述PYR的代谢机理提供有力证据。
3)蛋白编码基因的GO注释分析。GO是一个国际标准化的基因功能分类体系,提供了一套动态更新的标准词汇表来全面描述生物体中基因和基因产物的属性。通过对菌株Alteromonas sp. P7的GO数据库共注释到的基因共有2 850个,占总编码基因的69.87%。
同一个基因基于不同的分类标准,会有多种注释。由图8可见,参与生物过程的基因有2 688个,最多的是参与细胞氮化合物代谢和生物合成过程的基因,分别有992和963个,另外,参与小分子代谢过程的基因有732个,参与转运的基因有537个。拥有分子功能的基因有2 568个,其中参与离子整合过程的基因占比最大,有897个,与氧化还原酶活性有关的有402个,参与DNA整合的有331个,与裂解酶活性有关的有303个。与细胞组分有关的基因有909个,大部分基因与细胞体有关,共有897个,与胞内物质相关的有599个,与细胞质相关的有457个。
4) PYR降解基因的预测.在微生物代谢降解芳香族化合物时起作用的重要酶为双加氧酶,其原理是利用空气中的活性因子在芳香化合物的邻位或者间位发生氧化反应,能作用于苯系物、邻苯二酚以及联苯等物质。不同的双加氧酶对底物并没有专一的选择性,即同一种双加氧酶可以作用多种底物,如邻苯二酚双加氧酶不仅可以作用于邻苯二酚,而且还可以氧化甲苯。
本研究对交替单胞菌进行了全基因组测序,并将测序结果通过比对各种数据库进行基因功能注释。根据这些注释筛选出了可能的双加氧和酯酶等相关降解酶。表2中列出了这些酶蛋白的ID和序列长度。邻苯二酚代谢是降解天然或人工合成的芳香化合物的重要途径,在该途径中,邻苯二酚2,3-双加氧酶和羧基粘康酸内酯脱羧酶酶是主要作用酶,这些酶的编码基因存在于菌株P7染色体上。
-
1)从受船舶用油污染的海底沉积物中筛选分离出1株PYR降解菌P7,经鉴定为交替单胞菌属(Alteromonas sp.)。当PYR的初始浓度为20 mg·L−1时,Alteromonas sp. P7降解率为44.8%。
2)菌株P7降解PYR的最适pH为7~8,最适盐度在3.0%~3.5%,最适温度为30 ℃,在PYR为10 mg·L−1 时,菌株P7对PYR的降解率最高。
3) Alteromonas sp. P7降解PYR过程中产生的中间代谢产物包括水杨酸和邻苯二甲酸,说明Alteromonas sp. P7对PYR的降解存在邻苯二甲酸途径和水杨酸途径。而且Alteromonas sp. P7具有邻苯二酚2,3-双加氧酶活力,由此可以确定Alteromonas sp. P7是以邻苯二酚为中间代谢物对PYR进行降解。
4) Alteromonas sp. P7框架图测序及其编码基因蛋白的功能注释表明,其基因组序列长度4 597 467 bp,编码基因有8 164个,平均GC含量为44.26%。检索到含有编码芳香环断裂双加氧酶、邻苯二酚-2,3-双加氧酶、羧基粘康酸内酯脱羧酶等酶的基因。
海洋芘降解菌Alteromonas sp.P7的降解特性及基因组初步分析
Degradation characteristics and genome analysis of marine pyrene degrading bacteria Alteromonas sp. P7.
-
摘要: 以芘(PYR)为唯一碳源,在青岛受石油烃污染的码头的海底沉积物中分离到1株PYR海洋降解菌株P7,经过形态学观察、16S rRNA基因序列分析鉴定菌株P7属于交替单胞菌(Alteromonas)。该菌株在20 d内对PYR的降解率可达44.8%。经GC-MS鉴定分析发现,菌株P7降解芘的代谢产物中含有邻苯二甲酸和水杨酸,且细胞代谢过程中产生邻苯二酚2,3-双加氧酶,由此推测出Alteromonas sp. P7对芘的降解中间产物中含有邻苯二酚,且对芘的降解是通过邻苯二甲酸途径和水杨酸途径实现的。通过对分离得到的Alteromonas sp. P7进行测序,得到1条全长4 597 467 bp的基因组,对该基因序列注释,得到编码基因8 164个,平均GC含量为44.26%。重将测序结果与数据库比对并检索,菌株P7含有参与编码邻苯二酚2,3-双加氧酶的基因。Abstract: Using pyrene (PYR) as the sole carbon source, a Marine PYR degrading strain P7 was isolated from the seabed sediments of an oil-contaminated wharf in Qingdao. The strain P7 was identified as Alteromonas sp. by morphological observation and 16S rRNA gene sequence analysis. The degradation rate of PYR by strain P7 was 44.8% within 20 days, which was the first report of pyrene degradation by Alteromonas. Phthalic acid and salicylic acid were identified as the metabolites by GC/MS analysis, and catechol 2, 3-dioxygenase was produced during cell metabolism. Therefore, catechol was inferred in the intermediate products of pyrene degradation by Alteromonas sp. P7. Pyrene was degraded by both phthalic acid pathway and salicylic acid pathway. By sequencing the isolated Alteromonas sp. P7, a genome of 4 597 467 bp was obtained, and 8164 coding genes were determined by annotating the gene sequence, with an average GC content of 44.26%. After comparing the sequencing results with the database and retrieving them, a gene involved in the encoding of catechol 2, 3-dioxygenase was found in strain P7.
-
Key words:
- pyrene degrading bacteria /
- Alteromonas /
- metabolites /
- dioxygenase /
- genome
-

-
表 1 P7非编码RNA预测的数据统计
Table 1. Statistical analysis for non-coding RNA prediction of P7
非编码
RNA类型拷贝
数量序列平均
长度/bp序列总
长度/bp非编码 RNA 序列总长度
占基因组序列百分比/%5S rRNA 1 111 111 0.002 4 16S rRNA 1 1 529 1 529 0.033 3 23S rRNA 1 2 878 2 878 0.062 6 tRNA 59 78 4 629 0.100 7 ncRNA 24 167 4 030 0.087 6 表 2 PYR降解相关酶的预测
Table 2. Prediction of PYR degradation related enzymes
序列名称 序列长度 NR数据库注释 WP_105930780.1 213 ɑ-二酮戊酸双加氧酶AlkB WP_120959161.1 284 芳香环断裂双加氧酶 WP_120959905.1 357 4-羟基苯丙酮酸双加氧酶 APD86758.1 352 芳香环断裂双加氧酶ɑ亚基 HAS77371.1 446 苯甲酸盐1,2双加氧酶大亚基 HAS77370.1 164 苯甲酸盐1,2双加氧酶小亚基 WP_156087408.1 307 邻苯二酚-2,3-双加氧酶 WP_105929916.1 210 芳香酸脱羧酶 WP_105932022.1 183 羧基粘康酸内酯脱羧酶 -
[1] ZHU Q, TAO F, PAN Q. Fast and selective removal of oils from water surface via highly hydrophobic core-shell Fe2O3@C nanoparticles under magnetic field[J]. ACS Applied Materials and Interfaces, 2010, 2(11): 3141-3146. doi: 10.1021/am1006194 [2] MARTINEAU D, LAGACE A, BELAND P, et al. Pathology of stranded beluga whales (Delphinapterus leucas) from the St. Lawrence Estuary, Quebec, Canada[J]. Journal of General Plant Pahtology, 1988, 98(7): 287-311. [3] JIN H M, KIM J M, LEE H J. Alteromonas as a key agent of polycyclic aromatic hydrocarbon biodegradation in crude oil-contaminated coastal sediment[J]. Science and Technology, 2012, 46: 7731-7740. doi: 10.1021/es3018545 [4] FUCHS G, BOLL M, HEIDER J. Microbial degradation of aromatic compounds-from one strategy to four[J]. Nature Reviews Microbiology, 2011, 9(11): 803-816. doi: 10.1038/nrmicro2652 [5] LEISOLA M, MEUSSDOERFFER F. Production and identification of extracellular oxidases of Phanerochaete chrysosporium[J]. Journal of Biotechnology, 1985(6): 379-382. [6] OBRISTA D, ZIELINSKAA B, PERLINGER J A, et al. Accumulation of polyeyelie aromatie hydrocarbons (PAHs) and oxygenated PAHs (OPAHs) in organie and mineral soil horizons from four U. S. remote forests PAHs (OPAHs) in organie and mineral soil horizons from four U. S. remote forests[J]. Chemosphere, 2015, 134: 98-105. doi: 10.1016/j.chemosphere.2015.03.087 [7] CHAN K H, CHU W. The prediction of partitioning coefficients for chemicals causing environmental concern[J]. Science of the Total Environment[J]. 2000, 248(1): 1-10. [8] PRINCE R C. Bioremediation of marine oil spills[J]. Oil & Gas Science & Technology, 2004, 151(4): 495-512. [9] HEDLUND B P, STALEY J T. Isolation and characterization of Pseudo Alteromonas strains with divergent polycyclic aromatic hydrocarbon catabolic properties[J]. Environmental Microbiology. 2006, 8(1): 178-182. [10] DYKSTERHOUSE S E, GRAY J P, HERWIG R P, et al. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments[J]. International Journal of Systematic and Evolutionary Microbiology, 1995, 45(1): 116-123. [11] 王佳楠, 石妍云, 郑力燕, 等. 石油降解菌的分离鉴定及4株芽胞杆菌种间效应[J]. 环境科学, 2015, 36(6): 2245-2251. doi: 10.13227/j.hjkx.2015.06.044 [12] LI C H, WONG Y S, WANG H Y, et al. Anaerobic biodegradation of PAHs in mangrove sediment with amendment of NaHCO3[J]. Journal of Environmental Sciences, 2015, 30(4): 148-156. [13] YUAN S Y, CHANG B V. Anaerobic degradation of five polycyclic aromatic hydrocarbons from river sediment in Taiwan[J]. Jounal of Environmental Science and Health, Part B, 2007, 42(1): 63-69. doi: 10.1080/03601230601020860 [14] CHANG B V, CHANG I T, YUAN S Y. Anaerobic degradation of phenanthrene and pyrene in mangrove sediment[J]. Bulletin of Environmental Contamination and Toxicology, 2008, 80(2): 145-149. doi: 10.1007/s00128-007-9333-1 [15] ZHANG W, WANG H, ZHANG R, et al. Bacterial communities in PAH contaminated soils at an electronic-waste processing center in China[J]. Ecotoxicology, 2010, 19(1): 96-104. doi: 10.1007/s10646-009-0393-3 [16] SONG X H, XU Y, LI G M, et a1. Isolation, characterization of Rhodococcus sp. P14 capable of degrading high-molecular-weight polycyclic aromatic hydrocarbons and aliphatic hydrocarbons[J]. Marine Pollution Bulletin, 2011, 62(10): 2122-2128. doi: 10.1016/j.marpolbul.2011.07.013 [17] 张宏波, 林爱军, 刘爽, 等. 芘高效降解菌的分离鉴定及其降解特性研究[J]. 环境科学, 2010, 31(1): 243-248. doi: 10.13227/j.hjkx.2010.01.032 [18] 王蕾, 聂麦茜, 王志盈, 等. 一株芽胞杆菌和一株黄杆菌代谢芘的摄取方式解析[J]. 环境科学学报, 2009, 29(5): 924-929. doi: 10.3321/j.issn:0253-2468.2009.05.005 [19] CHEN Q, LI J, MEI L, et al. Study on the biodegradation of crude oil by free and immobilized bacterial consortium in marine environment[J]. Plos One, 2017, 12(3): 1-23. [20] 张建峰, 苏凤宜, 邢新会. 邻苯二酚2, 3-双加氧酶在恶臭假单胞杆菌整细胞催化中的酶活检测方法[J]. 化工学报, 2008(2): 450-455. doi: 10.3321/j.issn:0438-1157.2008.02.030 [21] 程晓宇, 刘伟伟, 许楹, 等. 中国东海和南海海域可培养烃类降解细菌的筛选及功能[J]. 微生物学通报, 2019, 46(5): 975-985. doi: 10.13344/j.microbiol.china.180685 [22] CAO B, MA T, REN Y, et al. Complete genome sequence of Pusillimonas sp. T7-7, a cold-tolerant diesel oil degrading bacterium isolated from the Bohai Sea in China[J]. Bacteriol. 2011, 193 (15): 4021-4022. [23] NZILA A, RAMIREZ C O, MUSA M M, SANKARA S, et al. Pyrene biodegradation and proteomic analysis in Achromobacter xylosoxidans, PY4 strain[J]. International Biodeterioration & Biodegradation. 2018, 130: 40-47. [24] SOHN J H, KWON K K, KANG J H, et al. Novosphingobium pentaromativorans sp. nov. , a high molecular mass polycyclic aromatic hydrocarbon degrading bacterium isolated from estuarine sediment[J]. International journal of systematic and evolutionary microbiology[J]. 2004, 54 (5), 1483-1487 [25] LYU Y, ZHENG W, ZHENG T, TIAN Y. Biodegradation of polycyclic aromatic hydrocarbons by Novosphingobium pentaromativorans US6-1[J]. PloS One 2014, 9(10): 1. [26] TONY H, AMEER K, RISKY K, et al. Biodegradation of pyrene by Candida sp. S1 under high salinity conditions[J]. Bioprocess & Biosystems Engineering, 2017, 40(9): 1411-1418. [27] MARTIN B C, GEORGE S J, PRICE C A, et al. The role of root exuded low molecular weight organic anions in facilitating petroleum hydrocarbon degradation: Current knowledge and future directions[J]. Science of the Total Environment, 2014, 472(15): 642-653. [28] REDA, BAYOUMI A, ASHRAF, et al. Optimization of bacterial biodegradation of toluene and phenol under different nutritional and environmental conditions[J]. Research Journal of Applied Sciences, 2010, 6(8): 1086-1095. [29] JAEKEL U, MUSAT N, ADAM B, et al. Anaerobic degradation of propane and butane by sulfate-reducing bacteria enriched from marine hydrocarbon cold seeps[J]. The ISME Journal, 2013, 7(5): 885-895. doi: 10.1038/ismej.2012.159 [30] XIE C J, FAN B, SUN Q Q, et al. Enhancement the enzymatic activity of phenol-degrading microbes immobilized on agricultural residues during the biodegradation of phenol in petrochemical wastewater[J]. Applied Mechanics and Materials, 2015, 737: 549-556. doi: 10.4028/www.scientific.net/AMM.737.549 [31] KAROLINA K, ANNA K, MAGDALENA W, et al. Biological inspirations: Iron complexes mimicking the catechol dioxygenases[J]. Materials, 2021, 14(12): 3250. doi: 10.3390/ma14123250 [32] KITA A, KITA S, FUJISAWA I, et al. An archetypical extradiol-cleaving catecholic dioxygenase: the crystal structure of catechol 2, 3-dioxygenase (metapyrocatechase) from Pseudomonas putida mt-2[J]. Structure, 1999, 7(1): 25-34. doi: 10.1016/S0969-2126(99)80006-9 [33] KOJIMA Y, ITADA N, HAYAISHI O. Metapyrocatechase: A new catechol-cleaving enzyme[J]. Journal of Biological Chemistry, 1961, 236(8): 2223-2228. doi: 10.1016/S0021-9258(18)64061-X -




 下载:
下载: