-
系统毒理学是建立在系统生物学基础上,综合多组学分析和传统毒理学方法,借助生物信息学[1-3]和计算毒理学[4-6]等模型化信息整合技术,对生物系统在外源化学物质扰动下保持稳定的能力进行评估,研究外源化学物质与生物系统相互作用机制的一门学科[7-9]。在还原论[10]思想引导下的毒理学,为研究某一种物质的毒性效应,只需要寻找并鉴别出与这一毒性效应直接相关的分子靶点即可。这是一种从整体到局部的简化研究思路。然而生物体信号通路的输入输出并不是由单一靶点控制的,而是由该通路的系统性结构和动力学共同控制的。有别于还原论思想,系统毒理学的核心思想是细胞和有机体等个体水平的形态和功能变化是由基因组、转录组和蛋白质组等系统扰动共同引起的。要理解化学物质在系统层次造成的生物学影响,就必须研究细胞和有机体整体的结构、功能特性和动力学机制,而非孤立部分的特征。系统毒理学分析框架强调系统性,可以减少跨物种外推、高剂量外推和低剂量外推的不确定性,有助于理解外源化合物对生命不同阶段的毒理作用以及遗传因素等对毒理作用的影响。
自21世纪初,组学技术为系统层次的计算建模奠定了数据基础[11]。从系统角度理解外源化学物质对生物的系统毒理学影响方面的研究逐渐增加[12-14]。如今多种组学技术的进步与组合,包括高精度的分子测量手段、高通量和高内涵的表征方法以及不断增强的计算能力、数据存储能力和信息管理工具,赋予了系统毒理学新的发展动力[15-17]。这些技术提供了海量的数据,也促使高质量数据库的建立[18].
组学数据可分为系统生物学数据和暴露相关数据两类,具有多维度、多尺度、彼此关联等特征。多维度是指整合的多组学数据描述了生物系统基因、蛋白质等不同维度的分子机制。多尺度是指组学数据既可表征生物系统的整体结构功能,又可用于描述局部的代谢通路。多组学数据彼此的关联性,体现在单一维度的组学研究无法解释某些生物表型现象[19-20]。
面对这些这些数据带来的发展机遇,美国国家研究咨询委员会编写了“21世纪毒性测试:远景与策略”这一研究报告。报告指出系统模型被期望建立以“毒性通路”为核心的量效关系模型,并基于给定的暴露条件将in vitro结果外推到in vivo人体血液和组织浓度[21]。为实现该目标,模型需要具备精准识别化学物质对生物系统扰动的能力。任何生物系统都由相互作用、相互依赖的若干组成部分结合而成,是具有特定功能的有机整体。这样的性质,可引导寻找能表示系统中各组分之间的相互作用关系的数学工具[22-23]。基于网络的模型有通用性强、灵活性强、包含节点间关系信息等优势,在系统毒理学中起到整合多组学数据、挖掘不同尺度的生物学信息的关键作用,得到了广泛应用[24-27]。
网络在数学上亦称图(Graph)[28]。基因调控和蛋白质相互作用等组学数据均可用网络表征。网络是节点与边的集合,网络中节点、边和网络整体结构3种信息可以共同用于描绘生物系统的结构功能。然而也正是这种灵活但不规则的数据结构对数据的处理和建模造成了很大阻碍。因此,以往系统毒理学对网络模型的分析方法局限于网络基本的拓扑学性质指标,例如节点的度(该节点连接的其他节点的数目)、节点的聚类系数(与该节点相邻的所有节点之间连边的数目占这些相邻节点之间最大可能连边数目的比例)等。已有较成熟的软件可以对网络图进行可视化与统计分析,如Cytoscape[29]、Pajek[30]。这种研究方式在慢性疾病防控、化学品风险评价等领域被应用[31-34]。然而,在人为选择用于描述网络的拓扑学性质指标时,网络中的部分信息被直接忽略,造成了信息损失。
近年来,信息科学领域开展了诸多针对网络的算法研究,可分为网络嵌入[35-36]与图神经网络(GNN)[37]。网络嵌入是一种将网络的结构特征整合进节点特征,进而实现网络特征化表示的信息挖掘手段。GNN则可以对大规模网络中的信息详尽地提取和学习[38],在基于网络的系统毒理学研究中有很大的应用潜力。其中,比较有代表性的算法是图卷积网络算法(GCN)[39]。GCN是一种深度学习算法,其优势在于能够从大规模网络数据中自动学习输入特征和输出决策之间的复杂关系,从而实现“端到端”的学习。换言之,在深度学习的特征提取过程中,模型不需要人为选择描述网络的拓扑指标作为模型的输入特征,而是可直接将用于描述生物系统的网络作为输入。基于此建立的模型,可充分利用网络中节点、边和网络结构3种信息,根据实际任务需求,综合完成如节点的分类预测、边的连接预测和网络图整体的分类预测等不同种类的预测任务。
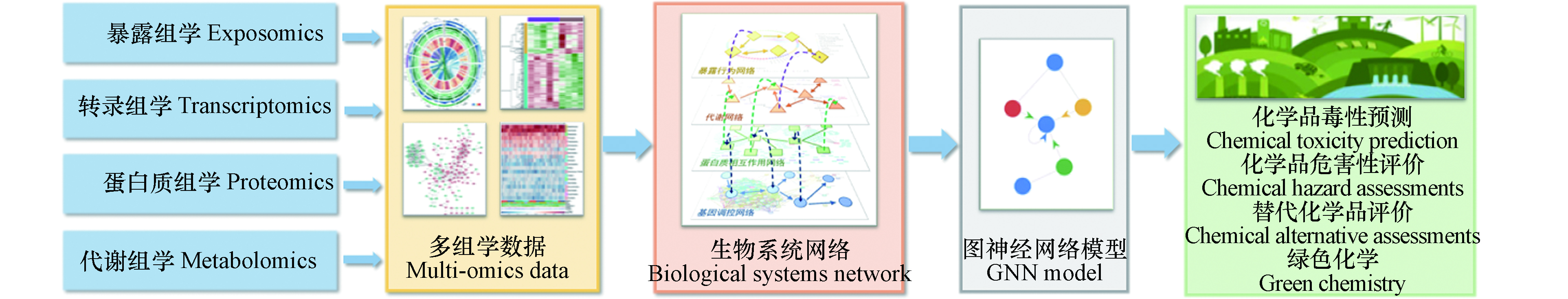
多组学数据的复杂性,使得以前的大多数研究仅仅侧重于分析单一类型的组学数据。组学间的相互作用尚未得到充分探讨[40]。基于网络的模型,可从多组学数据中提取数据间的逻辑关系。但传统网络分析方法在异质网络上的信息提取能力和计算效率较差[41]。因此,GNN模型有望被用于构建多维度、多尺度的先进系统毒理学模型,用于化学品有害效应的模拟预测(图1)。
本文介绍网络分析方法的研究策略、在系统毒理学领域中的应用,并对GNN模型在系统毒理学领域的应用进行展望。
-
基于网络的系统毒理学研究,可分为静态网络与动态网络。静态网络是对生物系统静态结构的表征,动态网络是对生物系统动态过程的模拟。动态网络往往在积累了足够的生物系统结构知识后,将研究聚焦于小规模的代谢通路,并通过微分方程组对生物系统的动态调控进行量化分析。例如,Schaff等[42]建立的真核细胞钙转运模型,专注于对真核细胞中钙动力学、钙离子流的模拟。此类动态网络无法反映出生物系统的整体结构特性。本文聚焦于静态网络的研究。
静态网络往往忽视化学物质在生物体内的动态过程,试图捕捉细胞或有机体在受到扰动后某一关键时刻内所有分子水平的特征,进而表征生物系统的结构与功能。静态网络的研究,通常包括网络构建与网络分析两环节。网络构建是指将复杂的生物系统抽象成节点与边组成的网络。节点被用来表示实体(Entity),在系统毒理学研究中通常为基因、蛋白质或化学物质等。边则通常被用来表示实体之间的逻辑关系,如基因之间的调控、蛋白质分子之间的相互作用、化学物质与蛋白质分子之间的相互作用等。
目前网络分析方法主要基于网络拓扑结构性质指标。例如,Karine等[33]基于化学品-蛋白质相互作用数据和基因表达数据构建网络模型,研究了农药DDT的异构体和代谢物,发现相比于代谢产物,母体DDT可能与更多疾病有关。Dai等[34]分别从肝毒性相关基因与化学物质的分子靶标角度出发,构建了肝毒性疾病模块网络与化学品-分子靶点相互作用网络,探究六溴环十二烷的肝毒性机制,并对其他环境污染物的肝毒性进行预测。
-
系统毒理学的目标,是得到一个可以模拟特定暴露情况下真实生物系统响应的理论模型,解释暴露与毒性效应之间的因果关系。实验技术与数学建模共同贯穿了这一目标的所有研究阶段,二者相互促进、相互依存[43]。对特定暴露情况下的生物系统进行大范围分子水平的测量,并综合分析多组学数据,就可以确定生物系统受暴露扰动的机制,有助于建立系统性的毒理学预测模型。
-
广义上,转录组学是指对细胞或有机体中所有RNA表达的变化的研究[44]。在生物系统研究中,转录组分析可以获得系统受暴露扰动后反应的全面机理解释,因此它是系统毒理学研究的重要方法。在大样本的转录组分析中,Langfelder等[45]开发的加权基因共表达网络分析(WGCNA)可用于对高度相关的基因进行模块化聚类,并使用模块中的关键基因来归纳模块特征,确定候选的生物标志或治疗靶点等。WGCNA促进了基于网络的基因筛查技术的发展,已应用于各种生物学领域,例如Tian等[46]利用WGCNA识别了与乳腺癌相关的核心模块和中心基因。WGCNA网络的构建,基于相关性系数(如皮尔逊相关系数、斯皮尔曼相关系数等),这就表示网络所能表示的关系只有相关性[47],无法说明因果性或其它更复杂的交互关系(例如三向交互)。
KEGG (Kyoto Encyclopedia of Genes and Genomes)是集合了基因和基因组高级功能的数据库[48]。基于KEGG数据库构建的大规模代谢网络可以在节点与边中包含更丰富的信息,使人们对观察到的生物表型有更深入的理解[49-51]。但由于缺乏对大规模网络信息挖掘的手段,基于KEGG网络的分析预测难以开展。IPA (Ingenuity Pathway Analysis)通路研究,可以利用大规模因果关系网络对上游的基因表达数据评分,并基于此预测生物功能和疾病等下游效应,实现对多组学数据结果的挖掘和分析预测[52]。近年来信息科学领域提出的网络数据结构、信息挖掘和分析预测手段,有望拓展这些网络模型的结构功能并改进其分析方式[53]。
-
与基因组、基因组学对应,Wilkins等[54]于20世纪90年代提出蛋白质组、蛋白质组学概念。蛋白质组是一个基因组、一种生物或一种细胞、组织所表达的全套蛋白质,蛋白质组学则致力于系统地描述蛋白质组中蛋白质子集的功能与性质。蛋白质组技术可通过识别一种外源化学物质在细胞的信号网络中发生作用的蛋白质靶标,来理解其影响信号通路活性的具体机理。Leonidas等[55]在利用基于生物化学数据建立的网络模型,揭示了原代肝细胞和转化型肝细胞之间 Toll样受体和炎症信号的差异。蛋白质相互作用网络也是蛋白质组学中常用的工具。在网络中处在核心地位的蛋白质,往往可以用于确定疾病基因与生物标志,但是连接较少的蛋白质同样可在生理过程中起到重要作用[56]。因此,开发多尺度的网络分析方法对网络中蕴含的细节信息进行深入挖掘是十分必要的。
-
代谢组学旨在对生物系统中的代谢物进行综合分析,提供关于生物系统的生化或生理状态变化以及化学物质引起的变化的详细信息,已成为系统生物学的一个重要组成部分。它可用于识别大量生物体内部的外源化学物质及其生物转化产物,或生物系统受扰动后产生的内源代谢物。二者共同组成的代谢组反映了暴露在基因组和蛋白质组维度上的分子扰动最终导致的代谢物质变化[57]。
然而,代谢组的数据分析仍然经常以还原论思想进行,即重视个别代谢物的变化。虽然这些方法有助于理解生物体代谢过程,但代谢关系的复杂本质未能得到有效探索。因此,许多基于网络的代谢组研究方法被相继提出,用于获取个体代谢物之间的差异、理解环境和遗传扰动的影响[58]。例如,Li等[59-60]为研究中药五味子对扑热息痛(对乙酰氨基酚)造成的肝脏毒性的抑制作用及其作用机制,构建了“化学物质-分子靶标-疾病”网络模型,同时考察了15条信号通路调控的52种相关蛋白。利用代谢组学和网络毒理学相结合的研究策略,揭示了化学物质导致的肝损伤的机制,并基于网络模型阐明了五味子治疗药物性肝损伤的作用机制。
代谢组学的研究融合了基因组学、转录组学以及蛋白质组学,以解释全面的暴露情况。与系统生物学不同,在系统毒理学的研究框架中,应重点关注外源物质暴露对生物系统造成的干扰。然而暴露行为复杂多变,亦有必要系统地对暴露行为展开研究,并以此将系统毒理学的研究框架拓展到环境暴露行为上。
-
暴露组是指从胎儿期开始,人的所有环境暴露(包括饮食、生活方式和感染等) [61]。Chakravarti等[62]指出基因和环境的共同作用是疾病最终结果的基础。Rappaport等[63]也指出,环境因素也是导致慢性疾病的主要原因。基于这些因素,Wild提出“我们迫切需要开发出与我们研究个体基因组同样精确的个体环境暴露方法”,用“暴露组”来匹配“基因组”以体现环境暴露研究的重要性[61]。然而,要实现如此大时间跨度的大规模测量,在方法学上存在很大挑战。
因此,暴露组研究仍主要为有针对性的、以生物标志物为基础的暴露测量,例如在某时间节点的一个或多个横断面的暴露测量[8]。这一情形与上文中静态网络的研究思路相契合:暂时忽略生化反应中的动态细节,而聚焦于系统整体的结构性质。此外,由于暴露组的研究对象极为复杂,环境相关性研究(EWAS)同样需要关注所研究体系中尽可能多的暴露相关性特征[64-65]。例如Robinson等[66]对孕期妇女进行了81种环境暴露行为的相关性分析,揭示了其中关键的暴露行为,指出未来的暴露研究应根据其与其他暴露的相关性来解释暴露效应。Vincent等[67]提出利用组学方法获得丰富的分子信息,将其用于关联疾病表型,进而研究广谱暴露(即外源性和内源性)与表型的关系。这将有希望发现新的环境风险因素,扩展对环境暴露与生物系统相互作用的理解。
-
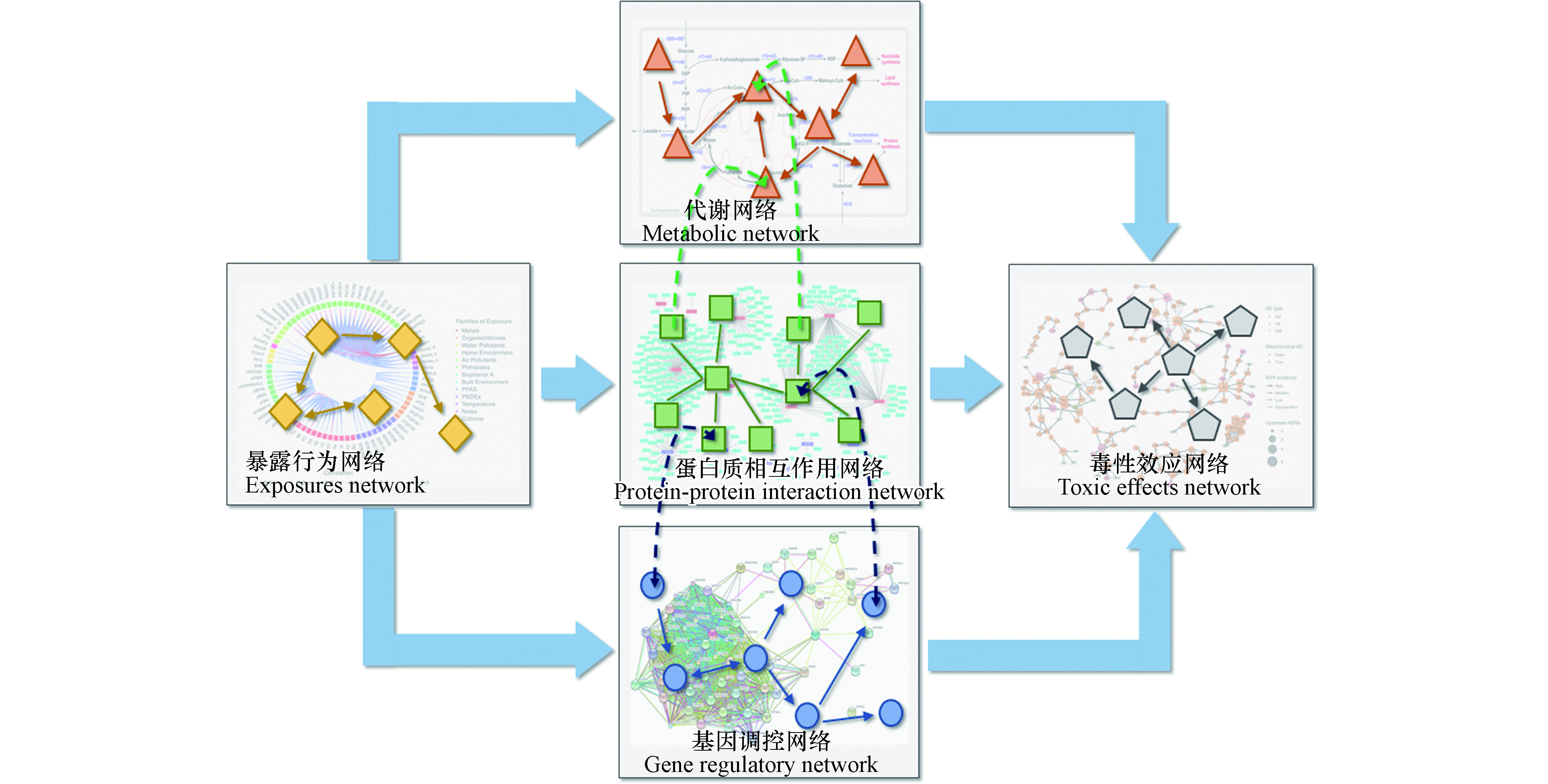
以往研究大多基于孤立的生物系统网络,如基因调控网络、蛋白质相互作用网络和化学物质-蛋白质作用网络等[50,55,59-60]。然而,系统毒理学的最终目标并不是分别在基因、RNA、蛋白质等系统内进行孤立研究,而是期望整合多组学数据,构建细胞或有机体整体的系统模型,以解释复杂的毒理效应[17]。如图2所示,多组学网络模型由不同生物系统网络模型整合而得,对多组学数据进行挖掘,捕获化学品暴露行为与毒性效应的内在联系。
大规模的网络可以表征丰富的逻辑关系和规则,网络的节点包含了可理解的特征标签,网络的边则包含了节点之间的依赖、从属、相似性等关系[68]。以预测某种内分泌干扰物的毒性效应为例,一个完备的系统毒理学模型不仅需要基于该物质所针对的分子靶点进行分析处理,还需要通过其毒性效应通路上下游之间的相互作用及调控行为进一步进行评估和预测。基于网络结构的模型有潜力利用不同维度生物系统中组成部件之间的交互作用关系,做出较为准确的关系预测。
针对该思路,已有研究利用异质网络(网络包含不同类型的节点与边,因此可以融合更多种类的实体和更复杂的逻辑关系)构建多种生物分子的复杂网络模型,并研究其结构与功能。例如,Liu等[69]构建了基因调控网络、蛋白质相互作用网络、代谢网络三层有相互关联的异质生物网络,并用数学方法定义了网络在受到扰动时的级联失效机制,数值模拟生物系统在扰动中保持自身功能稳定的能力。这一研究表征了每个基因对生物系统整体稳定性的贡献,也验证了在网络模型中融合不同维度生物系统间相互作用的可行性与重要性。
近年来信息科学领域开发了多种针对大规模复杂网络的分析方法,有望拓展网络模型的结构与功能,其中GNN模型展现出了良好的应用前景[40]。
-
针对网络拓扑结构性质的网络分析方法,在系统毒理学领域有较广泛的应用,但其数据挖掘和分析能力仍制约了基于网络分析方法的系统毒理学发展。尽管深度学习在过去10年中在很多领域取得成功[70-71],但它在毒理学和化学品风险评价中的应用仍有许多问题需要解决[72-73]。与传统人工智能建模方式利用的向量矩阵类数据相比,系统毒理学研究中网络数据结构可以表达更复杂的信息,然而也成为建模有待攻克的瓶颈[74-75]。很多研究针对上述问题,并开发了GNN模型,从算法角度给出了解决方案[76-79]。这里简述其中最具代表性的GCN算法[39]的基本原理,并对其在系统毒理学领域潜在的应用场景进行展望。
-
一种特殊结构的深层神经网络,卷积神经网络(CNN)[79]已经在结构化数据(例如图像)处理领域取得了成功。以图像处理为例,CNN作为一种深度学习模型,可以通过卷积算子从图像中自动提取与训练任务相关的特征,因此其在图像相关任务中表现极佳。
GCN将卷积算子的应用从结构化数据推广至非结构化数据(例如生物系统网络)。目前在知识图谱、分子图等图形领域应用较广的是空间GCN (S-GCN)。S-GCN的核心思想,是通过聚合相邻节点的特征来更新某一中心节点的特征表示,其具体手段是对网络图局部的邻居节点进行卷积操作。在邻居和中心节点之间的信息流动过程也称为消息传递(message passing)[75]。消息传递到中心节点的方式发展出了不同的传播规则,不同的规则以不同的方式整合网络的结构特征。
消息传递可以划分为两个阶段:消息传递阶段(message passing phase)和读出阶段(readout phase)。消息传递阶段如式(1)和(2)所描述:
式中,
$ {h}_{v} $ 和$ {h}_{w} $ 分别表示v和w两个节点的特征(例如在蛋白质相互作用网络中某一蛋白质分子的分子量),$ {e}_{vw} $ 表示连接节点对v和w的边的特征(例如在蛋白质相互作用网络中某一个蛋白质分子对的相互作用关系);$ {N}_{v} $ 表示节点v的一组邻居节点;$ {M}_{t}\left(\right) $ 表示将邻居节点特征传递到中心节点的消息传递函数。聚合了邻居节点的特征后,第t +1层中心节点的特征将通过更新函数$ {U}_{t}\left(\right) $ 更新。读出阶段利用读出函数
$ R\left(\right) $ ,根据最后一次消息传递后更新的所有节点特征$ {h}_{v}^{T} $ 为整个网络图$ G $ 计算特征向量$ \widehat{y} $ :根据消息传递函数
$ {M}_{t}\left(\right) $ 、更新函数$ {U}_{t}\left(\right) $ 、读出函数$ R\left(\right) $ 的选择,可以对模型进行变体以适应不同的任务需求。这种深度学习架构,实现了GCN模型对复杂网络“端到端”的学习过程,以避免人为筛选的特征无法完整描述网络的结构信息。此外,GCN还具有局部感知特征机制。局部感知是指某一层网络中的一个节点只与上一层中部分节点进行消息传递,在下一层网络中整合局部信息,从而获得网络的全局信息。这些特征机制,使GCN模型可以在关注网络局部重要信息的同时,对网络进行完整的学习,实现较好的模型效果。
-
系统毒理学研究中,重要的研究任务有:(1) 化学物质与蛋白质(分子靶标)之间的相互作用预测(分子起始事件[80-82]的预测),以判定化学物质是否会引起一系列毒性效应;(2) 蛋白质相互作用或基因调控关系预测,以探究化学物质毒性效应通路[83-84]的结构性质,或研究生物系统中的关键蛋白质或基因;(3) 化学物质与生物系统相互作用潜在的相关性预测,以探究复杂暴露行为之间是否相关联;(4) 生物系统网络不同尺度的结构性质研究,以揭示化学物质毒性效应的机制。
前述任务广义上可归纳为3类:生物系统网络中节点性质的研究、节点间逻辑关系的研究和网络结构性质的研究。这3类研究分别对应于GNN模型中的3种任务:节点的分类研究、边的连接预测研究、网络图的结构研究。前人已成功利用GCN算法,根据单细胞基因表达数据推断细胞之间的基因调控关系[53]。GNN模型的出现,为使用深度学习架构研究上述科学问题提供了一种新的方式。Zhang等[73]对当今较为流行的GNN模型进行了整理,提出了GNN模型几个关键的发展方向与挑战:(1)对异质网络的深度学习建模;(2) 对动态网络图的演化特征建模;(3) GNN模型的可解释性研究;(4)不同GNN模型架构间的组合性研究。2020年,整合了多组学信息和药物结构特征的异质图神经网络模型DeepCDR已经成功用于预测药物抗癌效能[40]。
综上,GNN是一个快速发展的领域,目前仍然存在挑战。但是,现有GNN模型框架已经可以为基于网络研究方法的系统毒理学研究注入新的动力。基于网络的分析方法,是系统毒理学研究中不可或缺的一部分,结合GNN对系统毒理学研究中的多组学数据建模,也是标志着系统毒理学进入深度学习时代的重要一步。
-
进入新世纪,毒理学的研究范式已经发生了重大转变。某些复杂且耗费大量资源的in vivo实验有望逐渐被in vitro测试和in silico模型所代替。然而这些研究方法,仍需在机理上展开更加深入的探究,降低跨物种外推、高剂量外推和低剂量外推带来的不确定性。以孤立的视角开展生物系统的毒理学研究,是一种过于简化的思想。基于图神经网络的系统毒理学,提供了新的方法与视角。其未来发展需要破解3方面的理论与技术瓶颈:(1) 系统毒理学建模所需数据的不完备性、网络模型中一些关键节点数据的缺失,可能会对网络的结构性质造成较大影响。(2) 基于静态生物网络的模型,忽略了生物系统动态的代谢过程。因此,需要对静态网络模型开展动态拓展研究,以定量描述化学物质在生物体内所导致扰动的时空变化。(3) 不同维度的生物网络间发生信息传递时,在一个维度上很小的不确定性,可能导致另一个维度上的预测出现很大的波动。
展望未来,系统毒理学赋予了网络模型更多的应用场景,快速发展的图神经网络也将助力系统毒理学的发展,实现更大规模的动态生物网络建模以及其对化学物质动态响应的模拟。目前,基于抽象的有害结局路径(AOP)的网络模型,已有望在化学品管控中发挥作用[85]。动态图神经网络的发展,则让网络模型除结构信息外也可以利用时序信息[86]。随着未来生物体不同维度网络模型的发展与整合,最终可能实现细胞或有机体整体的系统模型构建,进而实现对化学物质风险的快速、准确、综合的评估,助力化学品的风险管控。
基于图神经网络模型的系统毒理学研究展望
Prospect of systems toxicology research based on graph neural network model
-
摘要: 系统毒理学是建立在系统生物学基础上,综合多组学分析和传统毒理学方法,借助生物信息学和计算毒理学等模型化信息整合技术,对生物系统在外源化学物质扰动下保持稳定的能力进行评估,研究外源化学物质与生物系统相互作用机制的一门学科。转录组、蛋白质组、代谢组、暴露组等多组学数据,有多维度、多尺度、多关联的特征,为系统毒理学建模奠定了数据基础。如何利用计算建模,对多组学数据进行有效挖掘成为有待攻克的瓶颈。针对多组学数据的特点,基于网络的模型有着通用性强、灵活性强、包含节点间关系信息等优势,在系统毒理学中起到整合与挖掘多组学数据的关键作用。图神经网络(GNN)作为一种深度学习方法,在系统毒理学建模中展现了良好的应用前景。本文介绍了系统毒理学的研究目的、网络分析方法的研究策略,对GNN在系统毒理学领域的应用进行了展望。
-
关键词:
- 系统毒理学 /
- 多组学 /
- 网络分析方法 /
- 图神经网络(GNN)
Abstract: Systems toxicology can be regarded as an interdiscipline that is based on systems biology and combines multi-omics data analysis and classical toxicology methods. With information integration technologies such as bioinformatics and computational toxicology, systems toxicology can evaluate ability of biological systems to maintain stability under perturbation of xenobiotics and describe interactions of xenobiotics with biological systems. Transcriptomics, proteomics, metabolomics and exposomics data are multi-dimensional, multi-scaled and multi-correlational, and can lay a data foundation for systems toxicology modeling. It has become a technological bottleneck to use computational modeling for effective mining the multi-omics data. Network-based models exhibit advantages of high generality and flexibility, can characterize relationship information between nodes, and thus are suitable for dealing with multi-omics data, and can play a key role in integrating and mining multi-omics data in systems toxicology. Graph neural network (GNN) as a deep learning method, shows good application prospects in systems toxicology modeling. In this perspective, research orientations of systems toxicology and research strategies of network analysis methods are introduced, and application of GNN in systems toxicology is envisaged.-
Key words:
- systems toxicology /
- multi-omics /
- network analysis method /
- graph neural network (GNN)
-

-
[1] HAGEN J B. The origins of bioinformatics [J]. Nature Reviews Genetics, 2020, 1: 231-236. [2] KANEHISA M, BORK P. Bioinformatics in the post-sequence era [J]. Nature Genetics, 2003, 33: 305-310. doi: 10.1038/ng1109 [3] GAUTHIER J, VINCENT A T, CHARETTE S J, et al. A brief history of bioinformatics [J]. Briefings in Bioinformatics, 2019, 20(6): 1981-1996. doi: 10.1093/bib/bby063 [4] 陈景文, 王中钰, 傅志强. 环境计算化学与毒理学[M]. 北京: 科学出版社, 2018, 22-34. CHEN J W, WANG Z Y, FU Z Q. Computational chemistry and toxicology of the environment[M]. Beijing: Science Press, 2018, 22-34(in Chinese).
[5] 王中钰, 陈景文, 乔显亮, 等. 面向化学品风险评价的计算(预测)毒理学 [J]. 中国科学:化学, 2016, 46(2): 222-240. WANG Z Y, CHEN J W, QIAO X L, et al. Computational toxicology: oriented for chemicals risk assessment [J]. Science China Chemistry, 2016, 46(2): 222-240(in Chinese).
[6] KAVLOCK R, DIX D. Computational toxicology as implemented by the US EPA: Providing high throughput decision support tools for screening and assessment chemical exposure, hazard and risk [J]. Journal of Toxicology and Environmental Health, Part B:Critical Reviews, 2010, 13(2-4): 197-217. doi: 10.1080/10937404.2010.483935 [7] 王先良, 徐顺清. 系统毒理学及其应用 [J]. 生态毒理学报, 2006, 1(4): 289-294. WANG X L, XU S Q. Systems toxicology [J]. Asian Journal of Ecotoxicology, 2006, 1(4): 289-294(in Chinese).
[8] SHANA J S, ALAN R B, REX E F, et al. Systems toxicology: from basic research to risk assessment [J]. Chemical Research in Toxicology, 2014, 27(3): 314-329. doi: 10.1021/tx400410s [9] THOMAS H, REX E F, PAUL J, et al. Systems toxicology: real world applications and opportunities [J]. Chemical Research in Toxicology, 2017, 30(4): 870-882. doi: 10.1021/acs.chemrestox.7b00003 [10] 桂起权. 解读系统生物学: 还原论与整体论的综合 [J]. 自然辩证法通讯, 2015, 37(5): 219. GUI Q Q. Reading of systems biology: integration of reductionism and holism [J]. Journal of Dialectics of Nature, 2015, 37(5): 219(in Chinese).
[11] AARDEMA M J, MACGREGOR J T. Toxicology and genetic toxicology in the new era of 'Toxicogenomics': impact of '-Omics' technologies [J]. Mutation Research, 2002, 499(1): 13-25. doi: 10.1016/S0027-5107(01)00292-5 [12] IDEKER T, GALITSKI T, HOOD L. A new approach to decoding life: systems biology [J]. Annual Review of Genomics and Human Genetics, 2001, 2: 343-372. doi: 10.1146/annurev.genom.2.1.343 [13] WATERS M D, BOORMAN G, BUSHEL P, et al. Systems toxicology and the chemical effects in biological systems (CEBS) knowledge base [J]. Environmental Health Perspectives, 2003, 111(6): 811-824. doi: 10.1289/ehp.5971 [14] KITANO H. Systems biology: A brief overview [J]. Science, 2002, 295(5560): 1662-1664. doi: 10.1126/science.1069492 [15] WATERS M D, FOSTEL J M. Toxicogenomics and systems toxicology: aims and prospects [J]. Nature Reviews Genetics, 2004, 5(12): 936-948. doi: 10.1038/nrg1493 [16] HARTUNG T, VLIET E, JAWORSKA J, et al. Food for thought [J]. Systems Toxicology ALTEX, 2012, 29: 119-128. [17] PLANT N J. An introduction to systems toxicology [J]. Toxicology Research, 2015, 4: 9-22. doi: 10.1039/C4TX00058G [18] 李杰, 李柯佳, 张臣, 等. 计算系统毒理学: 形成、发展及应用 [J]. 科学通报, 2015, 60(19): 1751-1760. doi: 10.1360/N972014-01400 LI J, LI K J, ZHANG C, et al. Computational systems toxicology: Emergence, development and application [J]. Chinese Science Bulletin, 2015, 60(19): 1751-1760(in Chinese). doi: 10.1360/N972014-01400
[19] QUINN R A, MELNIK A V, VRBANAC A, et al. Global chemical effects of the microbiome include new bile-acid conjugations [J]. Nature, 2020, 579: 123-129. doi: 10.1038/s41586-020-2047-9 [20] KIMURA I, MIYAMOTO J, KITANO R O, et al. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice [J]. Science, 2020, 367(6481): 8429. doi: 10.1126/science.aaw8429 [21] KREWSKI D, JR. A D, ANDERSEN M, et al. Toxicity testing in the 21st century: A vision and a strategy [J]. Journal Toxicology Environmental Health-Part B-Critical Reviews, 2010, 13: 51-138. doi: 10.1080/10937404.2010.483176 [22] ZHANG Q, BHATTACHARYA S, ANDERSEN M E, et al. Computational systems biology and dose-response modeling in relation to new directions in toxicity testing [J]. Journal of Toxicology and Environmental Health, Part B:Critical Reviews, 2010, 13(2-4): 253-276. doi: 10.1080/10937404.2010.483943 [23] ZHANG Q, BHATTACHARYA S, CONOLLY R B, et al. Molecular signaling network motifs provide a mechanistic basis for cellular threshold responses [J]. Environmental Health Perspectives, 2014, 122(12): 61-70. [24] SHAO Z M, WANG K K, ZHANG S Y, et al. Ingenuity pathway analysis of differentially expressed genes involved in signaling pathways and molecular networks in RhoE gene-edited cardiomyocytes [J]. International Journal of Molecular Medicine, 2020, 46(3): 1225-1238. doi: 10.3892/ijmm.2020.4661 [25] KANEHISA M, GOTO S, FURUMICHI M, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs [J]. Nucleic Acids Research, 2010, 28(Suppl_1): 355-360. [26] ZHANG Y J, LIN H F, YANG Z H, et al. A method for predicting protein complex in dynamic PPI networks [J]. BMC Bioinformatics, 2016, 17(7): 229. [27] MANIPUR I, GRANATA I, MADDALENA L, et al. Clustering analysis of tumor metabolic networks [J]. BMC Bioinformatics, 2020, 21: 349. doi: 10.1186/s12859-020-03564-9 [28] BATTAGLIA P W, HAMRICK J B, BAPST V, et al. Relational inductive biases, deep learning, and graph networks [EB/OL]. [2021-5-15]. arXiv preprint, 2018,https://export.arxiv.org/pdf/1806.01261. [29] SHANNON P, MARKIEL A, OZIER O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks [J]. Genome Research, 2003, 13: 2498-2504. doi: 10.1101/gr.1239303 [30] BATAGELJ V, MRVAR A. Pajek-program for large network analysis [J]. Connections, 1998, 21: 47-57. [31] JAN S, ANNE C G. SnapShot: protein-protein interaction networks [J]. Cell, 2011, 144(6): 1000-1001. doi: 10.1016/j.cell.2011.02.025 [32] JOERG M, AMITABH S, MAKSIM K, et al. Uncovering disease-disease relationships through the incomplete interactome [J]. Science, 2015, 347(6224): 1257601. doi: 10.1126/science.1257601 [33] KARINE A, PHILIPPE G. Application of computational systems biology to explore environmental toxicity hazards [J]. Environmental Health Perspectives, 2011, 119(12): 1754-1759. doi: 10.1289/ehp.1103533 [34] DAI W N, TANG T T, DAI Z H, et al. Probing the mechanism of hepatotoxicity of hexabromocyclododecanes through toxicological network analysis [J]. Environmental Science & Technology, 2020, 54(23): 15235-15245. [35] GROVER A, LESKOVEC J. Node2vec: scalable feature learning for networks [EB/OL]. [2021-5-15]. arXiv preprint, 2016, https://cs.stanford.edu/people/jure/pubs/node2vec-kdd16.pdf. [36] FIGUEIREDO D R, RIBEIRO L F R, SAVERESE P H P. Struc2vec: Learning node representations from structural identity [EB/OL]. [2021-5-15]. arXiv preprint, 2017, https://export.arxiv.org/pdf/1704.03165. [37] SEJNOWSKI T J. The unreasonable effectiveness of deep learning in artificial intelligence[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(48): 30033-30038. [38] XU K, HU W, LESKOVEC J, et al. How powerful are graph neural networks [EB/OL]. [2021-5-15]. arXiv preprint, 2018, https://export.arxiv.org/pdf/1810.00826. [39] BRUNA J, ZAREMBA W, SZLAM A, et al. Spectral networks and locally connected networks on graphs [EB/OL]. [2021-5-15]. arXiv preprint, 2013,https://arxiv.org/pdf/1312.6203.pdf. [40] LIU Q, HU Z Q, JIANG R, et al. DeepCDR: A hybrid graph convolutional network for predicting cancer drug response [J]. Bioinformatics, 2020, 26(Supplement_2): 911-918. [41] ZHANG F, WANG M H, XI J N, et al. A novel heterogeneous network-based method for drug response prediction in cancer cell lines [J]. Scientific Reports, 2018, 8: 3355. doi: 10.1038/s41598-018-21622-4 [42] SCHAFF J, FINK C, SLEPCHENKO B, et al. A general computational framework for modeling cellular structure and function [J]. Biophysical Journal, 1997, 73(3): 1135-1146. doi: 10.1016/S0006-3495(97)78146-3 [43] YAHYA F A, HASHIM N F, ALI D A I, et al. A brief overview to systems biology in toxicology:The journey from in to vivo, in-vitro and -omics [J]. Journal of King Saud University-Science, 2020, 33(1): 101254. [44] KRISTIN S, BEAT B F, DANIELLE J M, et al. Transcriptomics in ecotoxicology [J]. Analytical and Bioanalytical Chemistry, 2010, 397(3): 917-923. doi: 10.1007/s00216-010-3662-3 [45] LANGFELDER P, HORVATH S. WGCNA: An package for weighted correlation network analysis [J]. BMC Bioinformatics, 2008, 9(559): 1471-2105. [46] TIAN Z L, HE W X, TANG J N, et al. Identification of important modules and biomarkers in breast cancer based on WGCNA [J]. OncoTargets and Therapy, 2020, 13: 6805-6817. doi: 10.2147/OTT.S258439 [47] 陈铭. 生物信息学(第3版)[M]. 北京: 科学出版社, 2018, 118-121. CHEN M. Bioinformatics (Third Edition) [M]. Beijing: Science Press, 2018, 118-121(in Chinese).
[48] KANEHISA M, FURUMICHI M, TANABE M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs [J]. Nucleic Acids Research, 2016, 45(D1): 353-361. [49] OBERHARDT M A, PUCHAIKA J, MARTINS V A P, et al. Reconciliation of genome-scale metabolic reconstructions for comparative systems analysis [J]. PLOS Computational Biology, 2011, 7(3): 1001116. doi: 10.1371/journal.pcbi.1001116 [50] PITKÄNEN E, JOUHTEN P, HOU J, et al. Comparative genome-scale reconstruction of gapless metabolic networks for present and ancestral species [J]. PLOS Computational Biology, 2014, 10(2): 1003465. doi: 10.1371/journal.pcbi.1003465 [51] KARLSEN E, SCHULZ C, ALMAAS E. Automated generation of genome-scale metabolic draft reconstructions based on KEGG [J]. BMC Bioinformatics, 2018, 19: 467. doi: 10.1186/s12859-018-2472-z [52] KRÄMER A, GREEN J, POLLARD J, et al. Causal analysis approaches in ingenuity pathway analysis [J]. Bioinformatics, 2014, 30(4): 523-530. doi: 10.1093/bioinformatics/btt703 [53] YUAN Y, JOSEPH Z B. GCNG: graph convolutional networks for inferring gene interaction from spatial transcriptomics data [J]. Genome Biology, 2020, 21: 200. doi: 10.1186/s13059-020-02088-y [54] WILKINS M R, SANCHEZ J C, GOOLEY A, et al. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it [J]. Biotechnology and Genetic Engineering Reviews, 1996, 13(1): 19-50. doi: 10.1080/02648725.1996.10647923 [55] LEONIDAS G A, JULIO S R, BENJAMIN D C, et al. Networks inferred from biochemical data reveal profound differences in toll-like receptor and inflammatory signaling between normal and transformed hepatocytes [J]. Molecular & Cellular Proteomics, 2010, 9(9): 1849-1865. [56] BOLTZ T A, DEVKOTA P, WUCHTY S. Collective influencers in protein interaction networks [J]. Scientific Reports, 2019, 9: 3948. doi: 10.1038/s41598-019-40410-2 [57] RAMIREZ T, DANESHIAN M, KAMP H, et al. Metabolomics in toxicology and preclinical research [J]. ALTEX-Alternatives to Animal Experimentation, 2013, 30(2): 209-225. [58] LEONARDO D S, SALEH A, YARIV B, et al. Network-based strategies in metabolomics data analysis and interpretation: from molecular networking to biological interpretation [J]. Expert Review of Proteomics, 2020, 17(4): 243-255. doi: 10.1080/14789450.2020.1766975 [59] LI X K, YANG H J, XIAO J C, et al. Network pharmacology-based investigation into the bioactive compounds and molecular mechanisms of schisandrae chinensis fructus against drug-induced liver injury [J]. Bio-organic Chemistry, 2020, 96: 103553. [60] LI X K, LI M Y, DENG S, et al. A network pharmacology-integrated metabolomics strategy for clarifying the action mechanisms of schisandrae chinensis fructus for treating drug-induced liver injury by acetaminophen [J]. Bio-organic & Medicinal Chemistry, 2021, 31: 115992. [61] WILD P C. Complementing the genome with an "Exposome":The outstanding challenge of environmental exposure measurement in molecular epidemiology [J]. Cancer Epidemiology Biomarkers & Prevention, 2005, 14(8): 1847-1850. [62] CHAKRAVARTI A, LITTLE P. Nature, nurture and human disease [J]. Nature, 2003, 421: 412-414. doi: 10.1038/nature01401 [63] RAPPAPORT S M. Discovering environmental causes of disease [J]. Journal of Epidemiology and Community Health, 2012, 66(2): 99-102. doi: 10.1136/jech-2011-200726 [64] KALLOO G, WELLENIUS G A, MCCANDLESS L, et al. Profiles and predictors of environmental chemical mixture exposure among pregnant women: the health outcomes and measures of the environment study [J]. Environmental Science & Technology, 2018, 52(17): 10104-10113. [65] CHEN H, ZHANG W X, ZHOU Y Q, et al. Characteristics of exposure to multiple environmental chemicals among pregnant women in Wuhan, China [J]. Science of the Total Environment, 2021, 754: 142167. doi: 10.1016/j.scitotenv.2020.142167 [66] ROBINSON O, BASAGANA X, AGIER L, et al. The pregnancy exposome: multiple environmental exposures in the inma-sabadell birth cohort [J]. Environmental Science & Technology, 2015, 49(17): 10632-10641. [67] VINCENT B, RUTHANN A R. Mapping the human exposome to uncover the causes of breast cancer [J]. International Journal of Environmental Research and Public Health, 2020, 17(1): 189. [68] RUIZ C, ZITNIK M, LESKOVEC J. Identification of disease treatment mechanisms through the multiscale interactome [J]. Nature Communications, 2021, 12: 1796. doi: 10.1038/s41467-021-21770-8 [69] LIU X M, ENRICO M, ARDA H, et al. Robustness and lethality in multilayer biological molecular networks [J]. Nature Communications, 2020, 11: 6043. doi: 10.1038/s41467-020-19841-3 [70] STEAD W. Clinical implications and challenges of artificial intelligence and deep learning [J]. JAMA, 2018, 320(11): 1107-1108. doi: 10.1001/jama.2018.11029 [71] SUN M Y, ZHAO S D, GILVARY C, et al. Graph convolutional networks for computational drug development and discovery [J]. Briefings in Bioinformatics, 2019, 21(3): 919-935. [72] TANG W, CHEN J W, WANG Z Y, et al. Deep learning for predicting toxicity of chemicals: A mini review [J]. Journal of Environmental Science and Health Part C- Environmental Carcinogenesis and Ecotoxicology Reviews, 2018, 36(4): 252-271. [73] ZHANG Z, CUI P, ZHU W. Deep learning on graphs: A survey [EB/OL]. [2021-5-15]. arXiv Preprint, 2018, https://export.arxiv.org/pdf/1812.04202. [74] ZHOU J, CUI G, ZHANG Z. Graph neural networks: A review of methods and applications [EB/OL]. [2021-5-15]. arXiv preprint, 2019, https://export.arxiv.org/ftp/arxiv/papers/1812/1812.08434.pdf. [75] GILMER J, SCHOENHOLZ S, RILEY P F, et al. Neural message passing for quantum chemistry [EB/OL]. [2021-5-15]. arXiv preprint, 2017, https://www.ics.uci.edu/~mohamadt/papers/Neural_message_passing.pdf. [76] HAMILTON W L, YING R, LESKOVEC J. Inductive representation learning on large graphs [EB/OL]. [2021-5-15]. arXiv preprint, 2017, https://cs.stanford.edu/people/jure/pubs/graphsage-nips17.pdf. [77] ATWOOD J, TOWSLEY D. Diffusion-convolutional neural networks [EB/OL]. [2021-5-15]. arXiv preprint, 2015, https://papers.nips.cc/paper/2016/file/390e982518a50e280d8e2b535462ec1f-Paper.pdf. [78] NIEPERT M, AHMED M, KUTZKOV K. Learning convolutional neural networks for graphs [EB/OL]. [2021-5-15]. arXiv preprint, 2016, http://proceedings.mlr.press/v48/niepert16.pdf. [79] KRIZHEVSKY A, SUTSKEVER L, HINTON G E. ImageNet classification with deep convolutional neural networks[C]. In Proceedings of the 25th International Conference on Neural Information Processing Systems, 2012, 1: 1097-1105. [80] ALLEN T E H, GOODMAN J M, GUTSELL S, et al. Defining molecular initiating events in the adverse outcome pathway framework for risk assessment [J]. Chemical Research in Toxicology, 2014, 27(12): 2100-2112. doi: 10.1021/tx500345j [81] ANKLEY G T, BENNETT R S, ERICKSON R J, et al. Adverse Outcome Pathways: A conceptual framework to support ecotoxicology research and risk assessment [J]. Environmental Toxicology and Chemistry, 2010, 29(3): 730-741. doi: 10.1002/etc.34 [82] KREWSKI D, ANDERSEN M E, TYSHENKO M G, et al. Toxicity testing in the 21st century: Progress in the past decade and future perspectives [J]. Archives of Toxicology, 2020, 94(1): 1-58. doi: 10.1007/s00204-019-02613-4 [83] CHEN S, ZHANG Z H, QING T, et al. Activation of the Nrf2 signaling pathway in Usnic Acid-induced toxicity in HepG2 cells [J]. Archives of Toxicology, 2017, 91: 1293-1307. doi: 10.1007/s00204-016-1775-y [84] LIU L, WU F Y, ZHU C Y, et al. Involvement of dopamine signaling pathway in neurodevelopmental toxicity induced by isoniazid in zebrafish [J]. Chemosphere, 2021, 265: 129109. doi: 10.1016/j.chemosphere.2020.129109 [85] DREIER D A, DANIELLE F M, JOEL N M, et al. Linking mitochondrial dysfunction to organismal and population health in the context of environmental pollutants: Progress and considerations for mitochondrial adverse outcome pathways [J]. Environmental Toxicology and Chemistry, 2019, 38(8): 1625-1634. [86] SKARDING J, GABRYS B, MUSIAL K. Foundations and modelling of dynamic networks using dynamic graph neural networks: A survey [EB/OL]. [2021-5-15]. arXiv preprint, 2020, https://export.arxiv.org/pdf/2005.07496. -



 下载:
下载: