-
镉是一种普遍的环境污染物,已被国际癌症研究机构列为一级致癌物[1 − 3]. 镉具有较高的迁移性,易被农作物吸收从而进入食物链,人体通过饮食和呼吸摄入等途径长期接触镉可对肾脏、骨骼和淋巴等组织器官造成不同程度的危害[4 − 5]. 镉是我国土壤的主要重金属污染物,工业和农业发展导致约7.0%的土壤镉浓度超标(0.3 mg·kg−1)[2, 6]. 因此,面对严峻的镉污染现状,对环境中的镉进行准确的检测至关重要. 目前,镉的检测方法主要包括分光光度法[7]、原子发射光谱法[8]、原子吸收光谱法[9]、电化学法[10]和质谱法[1]等物理化学分析方法[11 − 12]. 这些方法通常需要结合繁琐的样品前处理手段,设备较昂贵,操作复杂,样品检测成本较高. 此外,这些传统分析方法多集中于镉总量分析,无法准确评估环境中镉的生物有效性. 生物有效性又称生物可利用度,一般指污染物由结合态转化为自由态后被生物吸收,并在生物体内靶点富集产生不良影响[13 − 14]. 镉的环境行为和生态健康风险主要与其在环境介质中的生物有效性有关[15]. 因此,对环境中的镉进行生物有效性检测是开展其环境风险评估和污染治理的重要前提.
镉全细胞生物传感器(cadmium whole-cell biosensors, Cd-WCBs)以微生物为载体,利用微生物自然进化出的调节元件重组成新的模块化基因回路,可响应胞内Cd2+并输出随Cd2+浓度变化的信号,是一种简单的镉生物有效性检测方法[16 − 17]. 1993年,Corbisier等以cadC为识别元件,lux为报告元件首次设计了基于转录因子的Cd-WCBs[18]. 随着对微生物与镉相互作用机制认识的深入,新的镉转录调控因子(如ZntR和CadR)被发现,通过结合不同的报告元件(如gfp和mcherry)和底盘微生物(如Escherichia coli和Pseudomonas putida),目前已构建了多种Cd-WCBs[19 − 20]. 另外,多种基于合成生物学的优化策略已被用于提高Cd-WCBs的检测灵敏度和特异性,推动了其在实际环境中的应用[21 − 22].
本文介绍了Cd-WCBs的分类和设计原理,从传感元件、传感模块和底盘细胞等方面总结了基于转录因子的Cd-WCBs常用优化策略,综述了Cd-WCBs在镉生物有效性检测中的应用现状,并讨论了Cd-WCBs在实际应用中的主要问题和未来发展方向.
-
镉全细胞生物传感器分为非特异性识别和特异性识别. 非特异性Cd-WCBs分为两类:一类利用金属调控蛋白(如金属硫蛋白Mtt1和Mtt5)结合Cd2+,促使蛋白自身结构发生变化,影响其与传感质粒上对应启动子的结合,进而促进下游荧光蛋白编码基因的转录,产生荧光信号(图1a)[23];另一类非特异性Cd-WCBs是基于酶生物传感器原理构建,当Cd2+与一些酶结合后,会抑制或增强酶活性,促使一些带电物质的消耗或产生,通过检测电信号可监测Cd2+浓度. 该非特异性Cd-WCBs利用藻类或微生物自身产生的酶与金属离子结合的特性,将这些细胞固定在牛血清白蛋白膜上,进而固定在电极上,该传感器具有较高的灵敏度和稳定性(图1b)[24 − 25]. 然而,Mtt1和Mtt5等金属调控蛋白可结合多种金属离子,藻类和微生物中尚未发现与Cd2+特异结合的酶,因此这两类非特异性Cd-WCBs难以实现对Cd2+的特异检测.
特异性识别Cd-WCBs也分为两类. 一类是基于转录因子的Cd-WCBs,即由特异识别蛋白编码基因和报告基因组成的双组分Cd-WCBs[26]. 当特异性识别蛋白(如CadC、ZntA和CadR等)与Cd2+结合后蛋白构象发生变化,影响其与传感质粒上对应启动子的结合,改变启动子区域DNA的构象,促使RNA聚合酶对下游报告基因转录,产生可检测的光学或电化学信号(图1c),这种传感器多用于环境样品的检测中[16]. 另一类特异性识别的Cd-WCBs是基于荧光共振能量转移(fluorescence resonance energy transfer, FRET)原理,通过构建质粒载体,在细胞中表达含有供体和受体荧光团的特异性识别蛋白,该重组蛋白与Cd2+特异结合后三维构象发生变化,使供体荧光团的荧光激发受体荧光团,而供体荧光分子自身的荧光强度衰减,可通过检测受体荧光团的荧光强度检测Cd2+(图1d)[27 − 28]. 目前已开发的Cd-WCBs大多基于双组分传感原理(图1c),因此本文主要介绍双组分Cd-WCBs.
-
基于转录因子的特异性Cd-WCBs的核心是由报告元件和识别元件组成的传感元件. 其中,报告元件决定了传感器的信号输出方式和检测方法. Cd-WCBs使用的报告元件包括3类:产生显色物质的元件(如类胡萝卜素合成相关基因crtⅠ和β-半乳糖苷酶基因lacZ),可直接用肉眼观测[19, 29];利用酶促反应产生发光物质的元件(如荧光素酶基因lux和luc),可检测其化学发光[30 − 31];荧光蛋白编码基因(如绿色荧光蛋白基因gfp、红色荧光蛋白基因rfp和mcherry),可通过荧光法检测[32 − 34]. 识别元件直接决定了Cd-WCBs对Cd2+的检测灵敏度和特异性,目前双组分Cd-WCBs多采用两类识别蛋白,一类是来自ArsR/SmtB家族的CadC蛋白[35 − 36],另一类是来自MerR家族的ZntR和CadR蛋白[37 − 38].
CadC(ArsR/SmtB家族)具有两个金属结合位点,一个结合位点是由半胱氨酸配体组成的α3N位点,优先结合Cd2+、Pb2+、Zn2+和Bi3+;另一个结合位点是不含半胱氨酸配体的α5位点,可与Co2+和Zn2+结合[39]. 当胞内没有这些金属离子时,CadC与DNA上对应的启动子结合抑制下游基因的表达;在这些金属离子存在时,CadC可识别并与之结合,减弱CadC与启动子的结合从而激活下游基因的表达[36, 40]. 采用CadC构建的Cd-WCBs对Cd2+的特异性较差,但具有较高的灵敏度[34, 41].
ZntR和CadR(MerR家族)与Cd2+的结合方式类似于MerR与Hg2+的结合[42]. ZntR的N端具有高度保守的螺旋-旋转-螺旋结构,与对应启动子区域内20 bp的间隔区结合,C端的金属结合域包括4个半胱氨酸残基(Cys114、Cys115、Cys124和Cys19)和一个组氨酸(His119),该配位环境适合与Zn2+、Cd2+、Hg2+和Pb2+结合[43]. 当ZntR通过C端金属结合域与这些金属离子结合后可转化为激活态蛋白,并向下扭曲DNA,缩短-10与-35区之间的距离,从而引起启动子区域DNA的构象变化,使启动子转变为最佳结合态,促进下游基因转录[37, 44]. 以zntR为识别元件构建的传感器有较高的灵敏度,但会受Pb2+和Zn2+的干扰[33, 45 − 46]. CadR具有两种变构位点可与Cd2+结合,变构位点Ⅰ由3个半胱氨酸残基(Cys77、Cys112和Cys119)和一个天冬酰胺残基(Asn81)组成,变构位点Ⅱ由两个组氨酸(His87和His90)、1个谷氨酸(Glu62)和尾部区域的可变配体组成. 当Cd2+与变构位点Ⅰ结合后会使启动子部分扭曲,随着Cd2+进一步结合到变构位点Ⅱ,CadR稳定到最佳DNA结合构象,并转化为稳定激活态,此过程缩短了−10和−35区之间的距离,便于与RNA聚合酶相互作用,促进下游基因的转录[47]. CadR在没有Cd2+存在的情况下抑制自身的表达,而在Cd2+存在的情况下被诱导表达. 此外,CadR具有独特的富组氨酸的C末端延伸,有助于其与Cd2+的结合[38]. 基于以上协同结合机制,CadR可以很好地区分Cd2+与其他金属离子,是目前已知的对Cd2+特异性最高的识别蛋白,利用该蛋白构建的传感器不受0—200 µg·L−1范围内的其他离子干扰,但仍受高浓度的Hg2+(> 200 µg·L−1)、Pb2+(> 200 µg·L−1)和Zn2+(> 240 µg·L−1)的干扰[31, 48 − 49]. CadC与ZntR/CadR这两类识别蛋白已被用于开发Cd-WCBs,并应用于水和土壤等样品中镉的生物有效性检测. CadC是最早于金黄色葡萄球菌中发现的Cd2+转录调控因子,对Cd2+具有较高的灵敏度,但特异性较差[36]. 相较于CadR,ZntR具有较高的灵敏度,但特异性较差,CadR是目前已知的对Cd2+特异性最高的识别蛋白,但灵敏度较低. 这两类识别蛋白启动子的作用方式不同. 在没有金属离子的情况下,这两类蛋白均抑制对应启动子的表达;当识别到金属离子后,CadC会减弱与对应启动子的结合从而激活下游基因表达,ZntR/CadR则通过改变启动子区域DNA的构象并与之紧密结合从而促进下游基因表达[50].
-
目前开发的Cd-WCBs均存在以下两个共性问题. 首先,Cd2+与特异识别蛋白的结合较弱,导致Cd-WCBs的灵敏度较低. 在实验室条件下,传感器的灵敏度大多高于10 µg·L-1[20, 46, 51],极少数通过优化后可低于1 µg·L-1[31, 52]. 其次,现基于多个识别元件(如cadC、zntA和cadR)构建的Cd-WCBs易受其他金属离子干扰. 因此需利用合成生物学手段提高Cd-WCBs的灵敏度和特异性[53].
-
识别蛋白的定向改造是对识别蛋白的编码序列进行定点突变,构建灵敏度和特异性更高的Cd-WCBs(图2). 研究发现,通过将cadR的C端截断10或21个氨基酸,可有效降低传感器对Zn2+和Hg2+的响应,提高Cd-WCBs的特异性[48]. 但是,这部分氨基酸的缺失影响了CadR与Cd2+的结合,降低了该传感器的荧光信号,导致Cd-WCBs对Cd2+的灵敏度降低[48].
通过定向改造还可产生新的特异性识别蛋白。例如,利用寡核苷酸定向诱变技术,将随机突变引入MerR的关键金属结合域,可筛选得到在Hg2+诱导下抑制GFP的荧光但在Cd2+诱导下发荧光的突变体,使特异性识别Hg2+的MerR对Cd2+高度敏感[54]. 非定向改造主要通过定向进化来筛选突变型识别蛋白,以获得灵敏度和特异性更高的突变型Cd-WCBs,其突变位点随机(图2). 例如,通过易错PCR法生成生物传感器的突变体文库,进行流式细胞荧光分选后分离出性能更高的Cd-WCBs变种epCadR5,该突变型菌株对Cd2+的特异性相较于野生型提高了6.8倍,与Cd2+的亲和力提高了1.5倍[55].
-
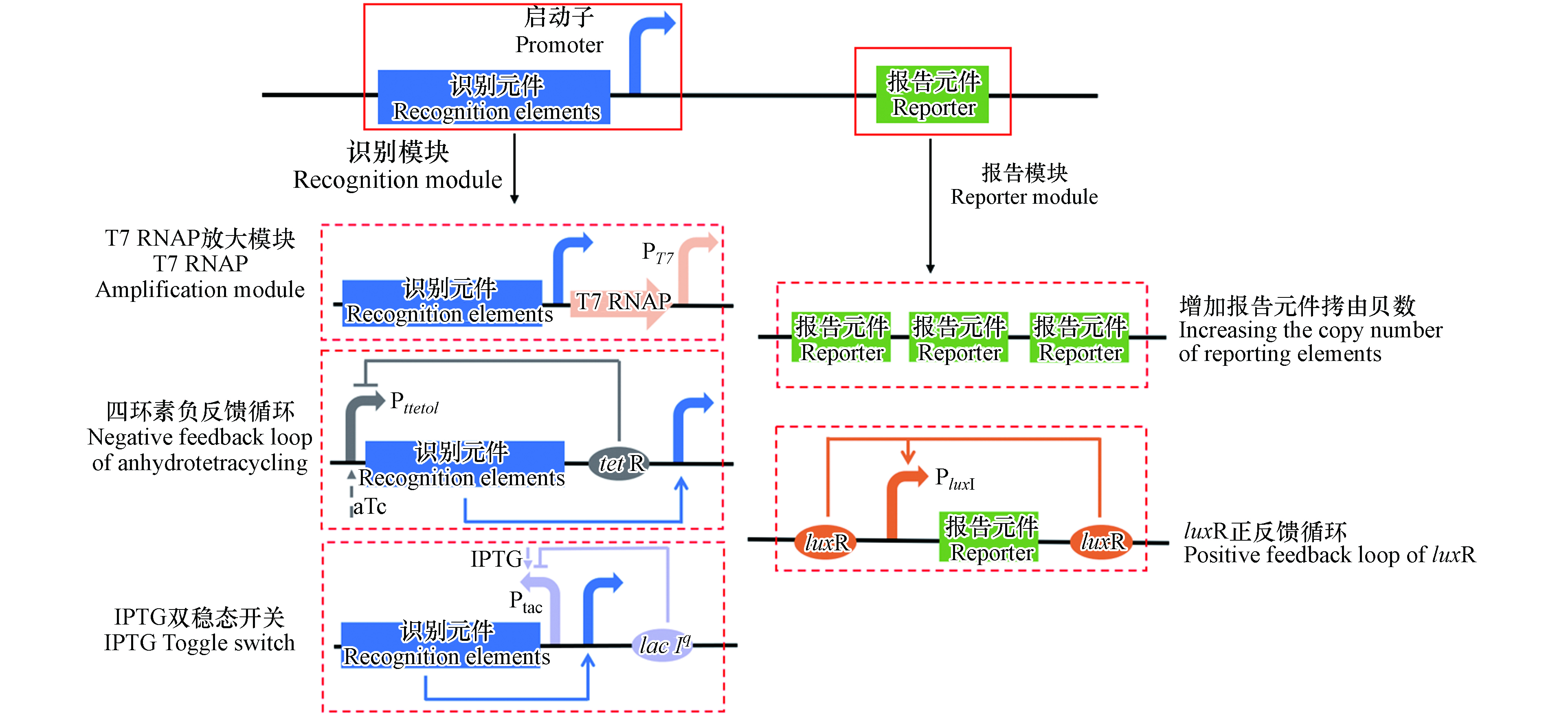
基因回路是将来自不同生物细胞的元件排列在传感网络中,通过不同的组合实现传感器的信号放大和传感回路切换等功能,提高传感器的灵敏度和特异性(图3). 例如,通过向传感元件中添加正反馈放大元件、增加报告元件拷贝数和添加T7RNAP扩增模块,可有效提高传感器的灵敏度和特异性,该Cd-WCBs对Cd2+的检测限由0.1 µmol·L−1降低至0.01 µmol·L−1,特异性提高了2.7倍[56]. 负反馈调控机制已被用于提高Cd-WCBs的灵敏度. Zhang等构建了一种受四环素调控的负反馈传感回路,在该传感回路中Plteto1调控cadR和tetR的表达,当添加四环素时,Plteto1启动子被激活,促进下游cadR和tetR的表达,而表达的TetR会抑制Plteto1启动子,形成负反馈回路[49]. 这种负反馈调控有效降低了胞内CadR的浓度,继而降低了传感器的背景信号,相较于没有负反馈调控的传感器,其灵敏度提高了400倍,同时对Cd2+表现出极高的特异性,是其他金属离子信号强度的17.3—41.4倍[49]. 利用Cd2+诱导的启动子PcadR和异丙基-b-D-硫半乳糖苷(IPTG)诱导的启动子Ptac构建双稳态开关也是一种负反馈调控. 当胞内存在Cd2+存在时,会与CadR结合导致PcadR被诱导,继而促进LacⅠ和GFP的生成,LacⅠ抑制Ptac启动子从而抑制cadR的表达,降低胞内CadR浓度;当添加IPTG时,Ptac被诱导促进cadR表达,产生的CadR又抑制PcadR启动子;通过添加不同浓度的IPTG可实现该传感器的回路切换,使全细胞生物传感器的检测限降低了20倍[57].
-
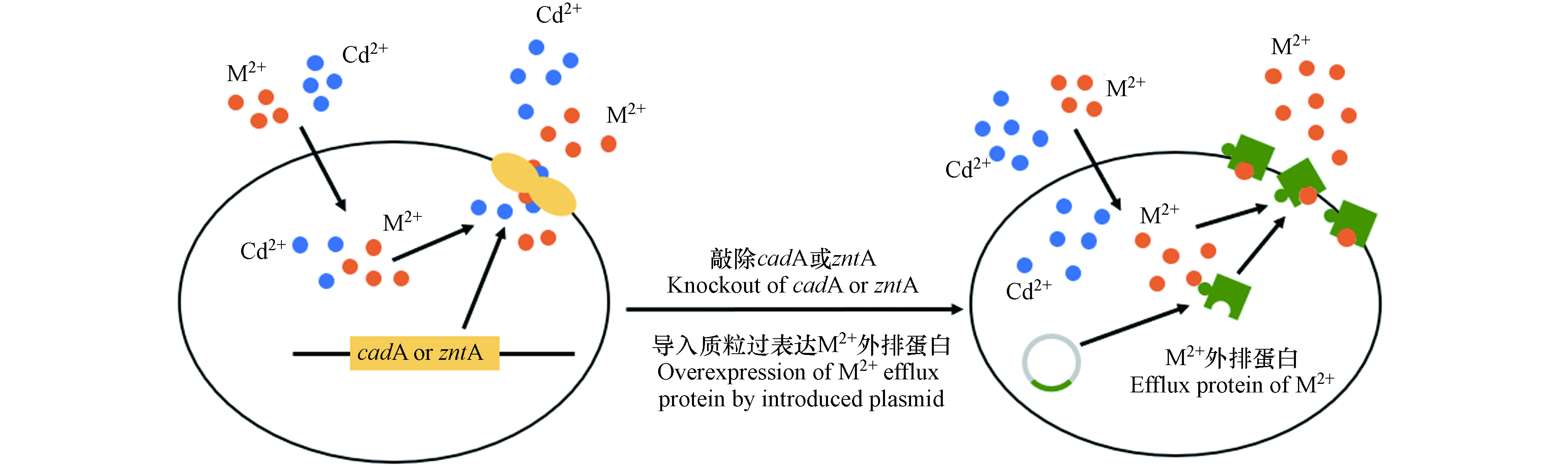
通过在不同菌株中导入相同的传感元件构建Cd-WCBs的灵敏度和特异性有较大差别[18, 20, 30],因此,底盘细胞对金属离子的调控也是影响Cd-WCBs灵敏度和特异性的重要因素. 全细胞生物传感器只能识别进入胞内的Cd2+,因此,通过对宿主菌的基因改造来提高Cd2+在胞内的积累[31, 58]和促进干扰离子的外排[52]是主要的改造策略(图4). 为了提高Cd2+在胞内的积累,可敲除宿主菌体内Cd2+相关外排蛋白编码基因(如Pseudomonas putida的cadA[31]和Escherichia coli的zntA[58]等)和二价离子外排通道编码基因[31, 52],从而将大部分Cd2+截留在胞内,提高传感器检测的灵敏度. 但是,金属离子外排蛋白的缺失可能会使干扰离子也被截留在胞内,因此还需配合其他手段来改善传感器的特异性,如过表达某些离子的外排蛋白编码基因(如Escherichia coli的Zn2+外排蛋白基因zitB)[52].
-
目前已开发出多种Cd-WCBs(表1),并应用于检测水和土壤等环境样品中镉的生物有效性. 大多Cd-WCBs采用大肠杆菌作为底盘细胞,识别元件以cadC、zntR和cadR为主,GFP是常用报告蛋白.
-
天然水体中镉的生物有效性主要受水样基质的影响. 在基质简单的天然水环境中,影响生物吸收利用镉的因素较少,传感器所检测到的镉浓度接近镉总量. 例如,利用一个受ZntR调控的PzntA启动子融合β-半乳糖苷酶编码基因lacZ构建Cd-WCBs,用于检测普通海水和淡水中不同浓度的Cd2+(5.6—112 µg·L−1),该传感器在海水与淡水中的检测结果一致[19]. 同样,对比去离子水和灌溉水,利用cadR和绿色荧光蛋白编码基因gfp构建Cd-WCBs的检测限以及线性范围几乎没有区别,在0—22.4 µg·L−1标准溶液中呈现较好的线性(R2灌溉水 = 0.995,R2去离子水 = 0.994)[52]. 然而,在富含悬浮颗粒物和有机质的天然水体中,受吸附和络合作用的影响,镉的生物有效性会降低,全细胞生物传感器检测到的Cd2+浓度低于总镉. 例如,以cadC和cadR为识别元件,egfp和mCherry为报告元件构建一种双传感系统全细胞生物传感器,用于检测不同来源水样中镉生物有效性,结果显示,未经处理的湖水会显著降低传感器的细胞密度和荧光信号,推测湖水中的悬浮颗粒物降低了镉的生物有效性[70].
-
土壤组成比天然水体更复杂,影响传感器活性和检测的因素较多,所以,Cd-WCBs一般用于土壤镉生物有效性的检测. 一般认为通过水浸提得到的水溶态Cd是可以被生物直接吸收利用的,然而,水溶态中含有的土壤胶体会吸附部分Cd2+,从而降低其生物有效性[73]. 因此,在测定沉积物和土壤水溶态Cd时,Cd-WCBs测得的Cd浓度比ICP-MS分析所得结果低,表明仅有部分水溶态Cd可被生物利用[41]. 此外,采用Cd-WCBs检测未过滤土壤-水悬浮液中的Cd浓度比水溶态Cd浓度高出20倍[46],表明土壤中部分与颗粒结合的Cd也可被生物吸收利用. Cd-WCBs还可用于评价土壤Cd污染的修复效率. 土壤淋洗是Cd污染的一种化学修复手段,在修复过程中常用螯合剂来提高淋洗效率. 利用Cd-WCBs对螯合剂淋洗修复前后的土壤中生物有效态Cd进行检测,发现淋洗修复虽然减少了土壤Cd的总量,但生物有效态Cd反而增加[33]. 这可能是因为淋洗过程中螯合剂破坏了Cd与土壤的结合,使部分土壤Cd由结合态转变为自由态,增加了土壤Cd的生物有效性[33]. 因此,Cd-WCBs具有评估土壤镉污染修复效率的应用前景.
-
Cd-WCBs还可直接用于检测一些基质简单的液体生物样品. 在Escherichia coli中利用cadC的对应启动子Pcad调控绿色荧光蛋白编码基因gfp的表达,所构建的全细胞生物传感器可在15 min内检测出牛奶中的Cd2+含量,检出限为10 µg·L−1,其检测结果与火焰原子吸收法一致[32]. 相较于火焰原子吸收法,Cd-WCBs对镉的检测无需样品预处理,检测速度更快,且具有高通量的优势. He等开发了一种可使用智能手机检测荧光的Cd-WCBs分析方法,该方法基于一种由cadR和gfp构建的Cd-WCBs,通过对Escherichia coli的金属调控系统进行基因编程,有效提高了传感器的特异性和灵敏度[52]. 该传感器可用于尿液中Cd2+的检测,其检测限低于世界卫生组织规定的安全标准,且在0—22.4 µg·L−1范围内有较好的线性(R2=0.981). 将Cd-WCBs置于样品中孵育8 h后离心收集,用蓝色LED板可激发Cd-WCBs的荧光,其荧光强度可被智能手机记录,最后利用ImageJ对图片中的荧光强度进行量化. 该方法成本低且使用方便[52].
-
Cd-WCBs可实现不同环境介质中镉的生物有效性检测,弥补了传统化学分析方法无法准确评估镉生物有效性的不足,有助于更好地揭示镉的环境归趋和评估其生态风险. 另外,与传统镉分析方法相比,Cd-WCBs具有使用方便、成本低廉、高通量和可实时检测的优势,因而具有良好的应用前景. 目前,Cd-WCBs的研究多集中对传感器灵敏度和特异性的优化,经不同策略优化后,Cd-WCBs检测限可低于欧盟允许的饮用水Cd浓度的最低安全标准(5 µg·L−1)[22, 29 − 30],并免受其他金属离子的干扰[39, 41, 43]. 然而,Cd-WCBs的实际应用仍有一定的局限性,通过以下几个方面工作可推动Cd-WCBs的实际应用.
(1)提高Cd-WCBs在不同环境介质中的存活率并维持其细胞活性是Cd-WCBs实际应用的关键. 作为Cd-WCBs的重要载体,底盘细胞易受环境因素(如温度、pH和营养源)影响,导致传感器的活性和功能受到影响. 另外,环境样品中高浓度Cd2+等其他金属对底盘微生物具有一定毒性,部分极端环境会抑制底盘微生物的生长,甚至导致细胞死亡. 因此,可通过开发新的底盘微生物或改造底盘微生物来提高其环境耐受能力. 此外,可构建双传感器系统Cd-WCBs来直观反映微生物存活状况[70].
(2)尽管实验室条件下已开发出多种高灵敏度和高特异性Cd-WCBs,但不同环境条件会影响Cd-WCBs的检测性能,可通过以下优化策略来提高Cd-WCBs在实际环境中的检测性能. 首先,可通过突变对识别元件进行改造或继续挖掘新的特异识别元件,以提高Cd-WCBs的特异性. 其次,通过不同生物元件之间的重组设计合理的基因回路,可进一步提高传感器的灵敏度和特异性. 比如,可通过增加放大模块、增加报告元件拷贝数、构建反馈循环或双稳态开关等手段提高Cd-WCBs的检测性能,利用逻辑门设计多输入传感器也是提高传感器特异性和灵敏度的一种可行手段[74]. 另外,底盘细胞的金属调控系统改造也是提高Cd-WCBs灵敏度和特异性的方法之一.
(3)目前,Cd-WCBs的原位和在线监测应用非常有限,这与其实际应用中可能导致的基因转移或释放有关. 许多国家和地区禁止转基因生物投入环境中,限制了Cd-WCBs在实际环境中的应用. 未来可利用Xeno核酸技术产生不被自然宿主机制识别的核酸序列、使用截止开关系统以及构建封闭的遏制装置等措施改造Cd-WCBs[75],来减少其遗传物质向土著微生物群落转移,进而有效推动Cd-WCBs在镉的环境监测与污染治理中的应用.
镉全细胞微生物传感器研究进展
Research progress of cadmium whole-cell biosensors
-
摘要: 镉全细胞微生物传感器(cadmium whole-cell biosensors,Cd-WCBs)以微生物为载体,利用微生物的调节元件组成模块化基因回路,以实现镉生物有效性的简单、经济和高通量检测. 本文概述了基于转录因子和酶生物传感器构建的非特异性Cd-WCBs,以及基于转录因子和荧光共振能量转移构建的特异性Cd-WCBs. 本文还总结了Cd-WCBs的基本优化策略,主要包括对识别蛋白进行定向改造或定向进化、利用反馈调控优化基因回路以及改造底盘细胞的金属调控系统. 目前,Cd-WCBs已经用于不同环境介质中镉的生物有效性检测,但是其在实际环境中细胞活性,和检测性能仍有待提高. 本研究指出未来可通过开发和改造底盘生物来提高传感器的环境适应能力,利用多种合成生物学手段进一步提高传感器的检测灵敏度和特异性.Abstract: Cadmium whole-cell biosensor (Cd-WCB), which contains a modular genetic circuit assembled by microbial regulation elements, can detect cadmium bioavailability in a simple, low-cost and high-through way. This study outlined the non-specific Cd-WCBs based on transcription factors and enzyme biosensors, and specific Cd-WCBs based on transcription factors and fluorescence resonance energy transfer. The optimization strategies of Cd-WCBs were summarized in this study, including directed mutagenesis and directed evolution of recognition proteins, optimization of genetic circuits with feedback loops, and modification of metalloregulatory system in chassis cells. Cd-WCBs have been applied for the assessment of cadmium bioavailability in different environmental samples, but the viability and detection performance in the actual environment need to be further improved. In future, chassis cells are suggested to be developed and modified to enhance the environmental adaptability of Cd-WCBs, and multiple synthetic biologic methods can be combined to improve the sensitivity and specificity of Cd-WCBs.
-
Key words:
- cadmium /
- whole-cell biosensor /
- bioavailability /
- environmental detection /
- specificity /
- sensitivity /
- recognition element /
- reporter.
-

-
表 1 已开发出的镉全细胞传感器
Table 1. Developed whole-cell biosensor for cadmium
调控蛋白基因
Regulatory protein gene底盘细胞
Chassis cells报告基因
Reporter gene检出限/(µg·L−1)
Limit of detection干扰离子
Interfering ions诱导时间/min
Induction time应用
Application参考文献
ReferencescadC Staphylococcus aureus lux 56 Bi3+、Pb2+ 120 — a [18] Escherichia coli lux 56 Bi3+、Pb2+ 60 — a Staphylococcus aureus luc 1.12 Pb2+、Sb3+ 120 — a [30] Bacillus subtilis luc 0.37 Sb3+、Zn2+、Sn4+ 120 — a [30] Escherichia coli gfp 10 Nb 15—30 牛奶 [32] Escherichia coli gfp 0.011 Pb2+、Sb3+ 120 沉积物、
土壤[41] Escherichia coli lacZ、rs-gfp 0.001 Pb2+、Zn2+ 120 — a [34] Staphylococcus aureus luc 100 Pb2+ 120 土壤 [59] Escherichia coli luc 1.12 Pb2+ 150 土壤 [60] — a gfp 56 Pb2+、Zn2+ 15 — a [61] Escherichia coli luc 11.2 — a 120 土壤 [62] zntR Escherichia coli gfp 11.2 Hg2+ 60 — a [58] Escherichia coli gfp 5 Hg2+、Zn2+ 960 — a [63] Escherichia coli lacZ 2.8 Nb 60 淡水、海水、土壤 [19] Escherichia coli lux 1.12 Hg2+、Pb2+、Zn2+ 60 — a [45] Escherichia coli egfp 500 Cr4+、Pb2+、Zn2+ 150 土壤 [33] Escherichia coli gfp 2 Hg2+、Zn2+ 90 — a [64] Escherichia coli gfp 1 Pb2+、Zn2+ 180 土壤 [65] Escherichia coli mcherry 200 Hg2+、Cu2+ 240 — a [66] Escherichia coli gfp 44.8 Pb2+、Zn2+ 120 — a [67] Escherichia coli luc 4.48 Hg2+、Zn2+ 120 土壤 [46] cadR Escherichia coli gfp 0.336 Nb 480 灌溉水、尿样 [52] Pseudomonas putida gfp 1.12 Nb 240 — a [57] Escherichia coli gfp 11.2 Hg2+、Zn2+ 120 — a [48] Escherichia coli gfp 112 Nb 840 — a [51] Escherichia coli gfp 0.45 Nb 120 河水、土壤 [55] Escherichia coli mcherry、bpsA、pcpS 5.48 Pb2+、Zn2+ 240 自来水、地表水 [68] Escherichia coli gfp、rfp 500 As3+、Hg2+、Pb2+ 240 地下水、海水 [20] Pseudomonas aeruginosa PAO-1 gfp、rfp 100 Hg2+、Pb2+ 240 地下水、海水 [20] Shewanella oneidensis MR-1 gfp、rfp 10000 Hg2+ 240 地下水、海水 [20] Enterobacter spp. NCR3 gfp、rfp 250 Hg2+、Pb2+ 240 地下水、海水 [20] Enterobacter spp. LCR17 gfp、rfp 1000 Hg2+、Pb2+ 240 地下水、海水 [20] Pseudomonas putida mcherry 1.12 Nb 360 — a [56] Pseudomonas putida mcherry 0.011 Nb 240 河水 [49] Pseudomonas putida lux 0.01 Pb2+、Zn2+ 180 — a [31] Pseudomonas putida rfp、egfp、lacZ 11.2 Hg2+ 1200 — a [69] cadC、cadR Escherichia coli egfp、mcherry 5.6 Nb 480 自来水、湖水 [70] cadR、merR Escherichia coli gfp、mcherry 11 Nb 480 — a [71] SEO1 Hansenula polymorpha gfp 112 Hg2+、As3+ 120 — a [72] crtⅠ Deinococcus radiodurans crtl 1.12 Nb — a — a [29] a. 文献未报导相关信息,以“—”表示;b. 该文献中Cd-WCBs在一定浓度范围内不受其他离子干扰.
a. “—” represents no relevant information was reported in the reference; b. Cd-WCBs in the reference are not disturbed by other ions in a certain concentration range. -
[1] 崔姗姗, 李占彬, 朱平, 等. 贵州遵义地区镉大气沉降通量与表层土壤分布特征[J]. 环境化学, 2022, 41(4): 1324-1334. doi: 10.7524/j.issn.0254-6108.2020122001 CUI S S, LI Z B, ZHU P, et al. Atmospheric deposition flux of cadmium and distribution characteristics of surface soil in Zunyi, Guizhou[J]. Environmental Chemistry, 2022, 41(4): 1324-1334 (in Chinese). doi: 10.7524/j.issn.0254-6108.2020122001
[2] HE S Y, HE Z L, YANG X E, et al. Soil biogeochemistry, plant physiology, and phytoremediation of cadmium-contaminated soils[J]. Advances in Agronomy, 2015, 134: 135-225. [3] International Agency for Research on Cancer (IARC). A review of human carcinogens-Part C: Arsenic, metals, fibres, and dusts [R]. Lyon, 2012. [4] EFSA) E F S A. Cadmium in food - Scientific opinion of the panel on contaminants in the food chain[J]. EFSA Journal, 2009, 7(3): 980. [5] SUHANI I, SAHAB S, SRIVASTAVA V, et al. Impact of cadmium pollution on food safety and human health[J]. Current Opinion in Toxicology, 2021, 27: 1-7. doi: 10.1016/j.cotox.2021.04.004 [6] 全国土壤污染状况调查公报[R]. 北京: 环境保护部, 2014. National soil contamination survey report[R]. Beijing: Ministry of Environmental Protection of China, 2014 (in Chinese) .
[7] SANTOS I C, MESQUITA R B R, RANGEL A O S S. Micro solid phase spectrophotometry in a sequential injection lab-on-valve platform for cadmium, zinc, and copper determination in freshwaters[J]. Analytica Chimica Acta, 2015, 891: 171-178. doi: 10.1016/j.aca.2015.08.021 [8] 陈晓晨, 黄艺佳, 赵桐, 等. 中国典型土壤中镉的生物可给性影响因素研究及其健康风险评估[J]. 环境化学, 2021, 40(10): 3015-3023. doi: 10.7524/j.issn.0254-6108.2021040204 CHEN X C, HUANG Y J, ZHAO T, et al. Influencing factors of Cd bioaccessibility in China’s representative soils and the human health risk assessment[J]. Environmental Chemistry, 2021, 40(10): 3015-3023 (in Chinese). doi: 10.7524/j.issn.0254-6108.2021040204
[9] 任杰, 刘晓文, 吴颖欣, 等. 不同尺度水稻土对镉的吸附解吸特征与定量分析研究[J]. 环境化学, 2020, 39(11): 3200-3212. doi: 10.7524/j.issn.0254-6108.2019082703 REN J, LIU X W, WU Y X, et al. Research of adsorption and desorption characteristics and quantitative analysis of cadmium in Paddy Soils with different scales[J]. Environmental Chemistry, 2020, 39(11): 3200-3212 (in Chinese). doi: 10.7524/j.issn.0254-6108.2019082703
[10] LI S, ZHANG C C, WANG S N, et al. Electrochemical microfluidics techniques for heavy metal ion detection[J]. The Analyst, 2018, 143(18): 4230-4246. doi: 10.1039/C8AN01067F [11] ROBARDS K, WORSFOLD P. Cadmium: Toxicology and analysis. A review[J]. The Analyst, 1991, 116(6): 549-568. doi: 10.1039/an9911600549 [12] WU P, LI C H, CHEN J B, et al. Determination of cadmium in biological samples: An update from 2006 to 2011[J]. Applied Spectroscopy Reviews, 2012, 47(5): 327-370. doi: 10.1080/05704928.2012.665401 [13] MCCARTY L S, MACKAY D. Enhancing ecotoxicological modeling and assessment. body residues and modes of toxic action[J]. Environmental Science & Technology, 1993, 27(9): 1718-1728. [14] LEE B G, LEE J S, LUOMA S N, et al. Influence of acid volatile sulfide and metal concentrations on metal bioavailability to marine invertebrates in contaminated sediments[J]. Environmental Science & Technology, 2000, 34(21): 4517-4523. [15] TANG X Y, ZHU Y G, CUI Y S, et al. The effect of ageing on the bioaccessibility and fractionation of cadmium in some typical soils of China[J]. Environment International, 2006, 32(5): 682-689. doi: 10.1016/j.envint.2006.03.003 [16] 侯启会, 马安周, 庄绪亮, 等. 微生物全细胞传感器在重金属生物可利用度监测中的研究进展[J]. 环境科学, 2013, 34(1): 347-356. doi: 10.13227/j.hjkx.2013.01.013 HOU Q H, MA A Z, ZHUANG X L, et al. Advance in the bioavailability monitoring of heavy metal based on microbial whole-cell sensor[J]. Environmental Science, 2013, 34(1): 347-356 (in Chinese). doi: 10.13227/j.hjkx.2013.01.013
[17] KIM H J, JEONG H, LEE S J. Synthetic biology for microbial heavy metal biosensors[J]. Analytical and Bioanalytical Chemistry, 2018, 410(4): 1191-1203. doi: 10.1007/s00216-017-0751-6 [18] CORBISIER P, JI G, NUYTS G, et al. luxAB gene fusions with the arsenic and cadmium resistance operons of Staphylococcus aureus plasmid PI258[J]. FEMS Microbiology Letters, 1993, 110(2): 231-238. doi: 10.1111/j.1574-6968.1993.tb06325.x [19] BIRAN I, BABAI R, LEVCOV K, et al. Online and in situ monitoring of environmental pollutants: Electrochemical biosensing of cadmium[J]. Environmental Microbiology, 2000, 2(3): 285-290. doi: 10.1046/j.1462-2920.2000.00103.x [20] BEREZA-MALCOLM L, ARACIC S, KANNAN R B, et al. Functional characterization of Gram-negative bacteria from different Genera as multiplex cadmium biosensors[J]. Biosensors and Bioelectronics, 2017, 94: 380-387. doi: 10.1016/j.bios.2017.03.029 [21] HUI C Y, GUO Y, LIU L S, et al. Recent advances in bacterial biosensing and bioremediation of cadmium pollution: A mini-review[J]. World Journal of Microbiology and Biotechnology, 2022, 38(1): 9. doi: 10.1007/s11274-021-03198-w [22] LIU C J, YU H, ZHANG B C, et al. Engineering whole-cell microbial biosensors: Design principles and applications in monitoring and treatment of heavy metals and organic pollutants[J]. Biotechnology Advances, 2022, 60: 108019. doi: 10.1016/j.biotechadv.2022.108019 [23] AMARO F, TURKEWITZ A P, MARTÍN-GONZÁLEZ A, et al. Whole-cell biosensors for detection of heavy metal ions in environmental samples based on metallothionein promoters from Tetrahymena thermophila[J]. Microbial Biotechnology, 2011, 4(4): 513-522. doi: 10.1111/j.1751-7915.2011.00252.x [24] CHOUTEAU C, DZYADEVYCH S, CHOVELON J M, et al. Development of novel conductometric biosensors based on immobilised whole cell Chlorella vulgaris microalgae[J]. Biosensors and Bioelectronics, 2004, 19(9): 1089-1096. doi: 10.1016/j.bios.2003.10.012 [25] HAN X J, LI C, YONG D M. Microbial electrode sensor for heavy-metal ions[J]. Sensors and Materials, 2019, 31(12): 4103. doi: 10.18494/SAM.2019.2645 [26] DELATOUR E, PAGNOUT C, ZAFFINO M L, et al. Comparative analysis of cell metabolic activity sensing by Escherichia coli rrnB P1- lux and Cd responsive- Lux biosensors: Time-resolved experiments and mechanistic modelling[J]. Biosensors, 2022, 12(9): 763. doi: 10.3390/bios12090763 [27] MIYAWAKI A, LLOPIS J, HEIM R, et al. Fluorescent indicators for Ca2+based on green fluorescent proteins and calmodulin[J]. Nature, 1997, 388(6645): 882-887. doi: 10.1038/42264 [28] CHIU T Y, CHEN P H, CHANG C L, et al. Live-cell dynamic sensing of Cd2+ with a FRET-based indicator[J]. PLOS ONE, 2013, 8(6): e65853. doi: 10.1371/journal.pone.0065853 [29] JOE M H, LEE K H, LIM S Y, et al. Pigment-based whole-cell biosensor system for cadmium detection using genetically engineered Deinococcus radiodurans[J]. Bioprocess and Biosystems Engineering, 2012, 35(1): 265-272. [30] TAURIAINEN S, KARP M, CHANG W, et al. Luminescent bacterial sensor for cadmium and lead[J]. Biosensors and Bioelectronics, 1998, 13(9): 931-938. doi: 10.1016/S0956-5663(98)00027-X [31] HYNNINEN A, TÕNISMANN K, VIRTA M. Improving the sensitivity of bacterial bioreporters for heavy metals[J]. Bioengineered Bugs, 2010, 1(2): 132-138. doi: 10.4161/bbug.1.2.10902 [32] KUMAR S, VERMA N, SINGH A K. Development of cadmium specific recombinant biosensor and its application in milk samples[J]. Sensors and Actuators B: Chemical, 2017, 240: 248-254. doi: 10.1016/j.snb.2016.08.160 [33] YOON Y, KIM S, CHAE Y, et al. Use of tunable whole-cell bioreporters to assess bioavailable cadmium and remediation performance in soils[J]. PLOS ONE, 2016, 11(5): e0154506. doi: 10.1371/journal.pone.0154506 [34] SHETTY R S, DEO S K, SHAH P, et al. Luminescence-based whole-cell-sensing systems for cadmium and lead using genetically engineered bacteria[J]. Analytical and Bioanalytical Chemistry, 2003, 376(1): 11-17. doi: 10.1007/s00216-003-1862-9 [35] LEBRUN M, AUDURIER A, COSSART P. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are present on Tn5422, a novel transposon closely related to Tn917[J]. Journal of Bacteriology, 1994, 176(10): 3049-3061. doi: 10.1128/jb.176.10.3049-3061.1994 [36] NUCIFORA G, CHU L, MISRA T K, et al. Cadmium resistance from Staphylococcus aureus plasmid PI258 cadA gene results from a cadmium-efflux ATPase[J]. Proceedings of the National Academy of Sciences of the United States of America, 1989, 86(10): 3544-3548. [37] OUTTEN C E, OUTTEN F W, O'HALLORAN T V. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli[J]. The Journal of Biological Chemistry, 1999, 274(53): 37517-37524. doi: 10.1074/jbc.274.53.37517 [38] LEE S W, GLICKMANN E, COOKSEY D A. Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator[J]. Applied and Environmental Microbiology, 2001, 67(4): 1437-1444. doi: 10.1128/AEM.67.4.1437-1444.2001 [39] BUSENLEHNER L S, WENG T C, PENNER-HAHN J E, et al. Elucidation of primary (α3N) and vestigial (α5) heavy metal-binding sites in Staphylococcus aureus PI258 CadC: Evolutionary implications for metal ion selectivity of ArsR/SmtB metal sensor proteins[J]. Journal of Molecular Biology, 2002, 319(3): 685-701. doi: 10.1016/S0022-2836(02)00299-1 [40] BUSENLEHNER L S, PENNELLA M A, GIEDROC D P. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance[J]. FEMS Microbiology Reviews, 2003, 27(2/3): 131-143. [41] LIAO V H C, CHIEN M T, TSENG Y Y, et al. Assessment of heavy metal bioavailability in contaminated sediments and soils using green fluorescent protein-based bacterial biosensors[J]. Environmental Pollution, 2006, 142(1): 17-23. doi: 10.1016/j.envpol.2005.09.021 [42] PERMINA E, KAZAKOV A, KALININA O, et al. Comparative genomics of regulation of heavy metal resistance in Eubacteria[J]. BMC Microbiology, 2006, 6(1): 49. doi: 10.1186/1471-2180-6-49 [43] CHANGELA A, CHEN K, XUE Y, et al. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR[J]. Science, 2003, 301(5638): 1383-1387. doi: 10.1126/science.1085950 [44] BROCKLEHURST K R, HOBMAN J L, LAWLEY B, et al. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli[J]. Molecular Microbiology, 1999, 31(3): 893-902. doi: 10.1046/j.1365-2958.1999.01229.x [45] RIETHER K, -A DOLLARD M, BILLARD P. Assessment of heavy metal bioavailability using Escherichia coli zntAp: Lux and copAp: Lux-based biosensors[J]. Applied Microbiology and Biotechnology, 2001, 57(5): 712-716. [46] IVASK A, VIRTA M, KAHRU A. Construction and use of specific luminescent recombinant bacterial sensors for the assessment of bioavailable fraction of cadmium, zinc, mercury and chromium in the soil[J]. Soil Biology and Biochemistry, 2002, 34(10): 1439-1447. doi: 10.1016/S0038-0717(02)00088-3 [47] LIU X C, HU Q Y, YANG J M, et al. Selective cadmium regulation mediated by a cooperative binding mechanism in CadR[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(41): 20398-20403. [48] TAO H C, PENG Z W, LI P S, et al. Optimizing cadmium and mercury specificity of CadR-based E. coli biosensors by redesign of CadR[J]. Biotechnology Letters, 2013, 35(8): 1253-1258. doi: 10.1007/s10529-013-1216-4 [49] ZHANG G B, HU S T, JIA X Q. Highly sensitive whole-cell biosensor for cadmium detection based on a negative feedback circuit[J]. Frontiers in Bioengineering and Biotechnology, 2021, 9: 799781. doi: 10.3389/fbioe.2021.799781 [50] BAKSH K A, ZAMBLE D B. Allosteric control of metal-responsive transcriptional regulators in bacteria[J]. Journal of Biological Chemistry, 2020, 295(6): 1673-1684. doi: 10.1074/jbc.REV119.011444 [51] AKBOĞA D, SALTEPE B, BOZKURT E U, et al. A recombinase-based genetic circuit for heavy metal monitoring[J]. Biosensors, 2022, 12(2): 122. doi: 10.3390/bios12020122 [52] HE M Y, LIN Y J, KAO Y L, et al. Sensitive and specific cadmium biosensor developed by reconfiguring metal transport and leveraging natural gene repositories[J]. ACS Sensors, 2021, 6(3): 995-1002. doi: 10.1021/acssensors.0c02204 [53] SOMAYAJI A, SARKAR S, BALASUBRAMANIAM S, et al. Synthetic biology techniques to tackle heavy metal pollution and poisoning[J]. Synthetic and Systems Biotechnology, 2022, 7(3): 841-846. doi: 10.1016/j.synbio.2022.04.007 [54] HAKKILA K M, NIKANDER P A, JUNTTILA S M, et al. Cd-specific mutants of mercury-sensing regulatory protein MerR, generated by directed evolution[J]. Applied and Environmental Microbiology, 2011, 77(17): 6215-6224. doi: 10.1128/AEM.00662-11 [55] CAI Y S, ZHU K L, SHEN L, et al. Evolved biosensor with high sensitivity and specificity for measuring cadmium in actual environmental samples[J]. Environmental Science & Technology, 2022, 56(14): 10062-10071. [56] JIA X Q, LIU T, MA Y B, et al. Construction of cadmium whole-cell biosensors and circuit amplification[J]. Applied Microbiology and Biotechnology, 2021, 105(13): 5689-5699. doi: 10.1007/s00253-021-11403-x [57] WU C H, LE D, MULCHANDANI A, et al. Optimization of a whole-cell cadmium sensor with a toggle gene circuit[J]. Biotechnology Progress, 2009, 25(3): 898-903. doi: 10.1002/btpr.203 [58] YOON Y, KANG Y, LEE W, et al. Modulating the properties of metal-sensing whole-cell bioreporters by interfering with Escherichia coli metal homeostasis[J]. Journal of Microbiology and Biotechnology, 2018, 28(2): 323-329. doi: 10.4014/jmb.1710.10012 [59] IVASK A, FRANÇOIS M, KAHRU A, et al. Recombinant luminescent bacterial sensors for the measurement of bioavailability of cadmium and lead in soils polluted by metal smelters[J]. Chemosphere, 2004, 55(2): 147-156. doi: 10.1016/j.chemosphere.2003.10.064 [60] HOU Q H, MA A Z, WANG T, et al. Detection of bioavailable cadmium, lead, and arsenic in polluted soil by tailored multiple Escherichia coli whole-cell sensor set[J]. Analytical and Bioanalytical Chemistry, 2015, 407(22): 6865-6871. doi: 10.1007/s00216-015-8830-z [61] BEABOUT K, BERNHARDS C B, THAKUR M, et al. Optimization of heavy metal sensors based on transcription factors and cell-free expression systems[J]. ACS Synthetic Biology, 2021, 10(11): 3040-3054. doi: 10.1021/acssynbio.1c00331 [62] HOU Q H, MA A Z, LI Y, et al. Assessing the effect of phosphate and silicate on Cd bioavailability in soil using an Escherichia coli cadAp: Luc-based whole-cell sensor[J]. Environmental Science. Processes & Impacts, 2014, 16(4): 890-896. [63] GIREESH-BABU P, CHAUDHARI A. Development of a broad-spectrum fluorescent heavy metal bacterial biosensor[J]. Molecular Biology Reports, 2012, 39(12): 11225-11229. doi: 10.1007/s11033-012-2033-x [64] ELCIN E, ÖKTEM H A. Inorganic cadmium detection using a fluorescent whole-cell bacterial bioreporter[J]. Analytical Letters, 2020, 53(17): 2715-2733. doi: 10.1080/00032719.2020.1755867 [65] HURDEBISE Q, TARAYRE C, FISCHER C, et al. Determination of zinc, cadmium and lead bioavailability in contaminated soils at the single-cell level by a combination of whole-cell biosensors and flow cytometry[J]. Sensors, 2015, 15(4): 8981-8999. doi: 10.3390/s150408981 [66] KIM Y, CHOI H, SHIN W H, et al. Development of colorimetric whole-cell biosensor for detection of heavy metals in environment for public health[J]. International Journal of Environmental Research and Public Health, 2021, 18(23): 12721. doi: 10.3390/ijerph182312721 [67] ZHANG C Y, SIDDIQUI S, NAVARRETE P M, et al. An integrated whole-cell detection platform for heavy metal ions[J]. IEEE Sensors Journal, 2020, 20(9): 4959-4967. doi: 10.1109/JSEN.2020.2964642 [68] HUI C Y, GUO Y, GAO C X, et al. A tailored indigoidine-based whole-cell biosensor for detecting toxic cadmium in environmental water samples[J]. Environmental Technology & Innovation, 2022, 27: 102511. [69] GUO Y, HUI C Y, ZHANG N X, et al. Development of cadmium multiple-signal biosensing and bioadsorption systems based on artificial Cad operons[J]. Frontiers in Bioengineering and Biotechnology, 2021, 9: 585617. doi: 10.3389/fbioe.2021.585617 [70] HUI C Y, GUO Y, WU J, et al. Detection of bioavailable cadmium by double-color fluorescence based on a dual-sensing bioreporter system[J]. Frontiers in Microbiology, 2021, 12: 696195. doi: 10.3389/fmicb.2021.696195 [71] HUI C Y, GUO Y, LI H, et al. Differential detection of bioavailable mercury and cadmium based on a robust dual-sensing bacterial biosensor[J]. Frontiers in Microbiology, 2022, 13: 846524. doi: 10.3389/fmicb.2022.846524 [72] PARK J N, SOHN M J, OH D B, et al. Identification of the cadmium-inducible Hansenula polymorpha SEO1 gene promoter by transcriptome analysis and its application to whole-cell heavy-metal detection systems[J]. Applied and Environmental Microbiology, 2007, 73(19): 5990-6000. doi: 10.1128/AEM.00863-07 [73] ABDU N, ABDULLAHI A A, ABDULKADIR A. Heavy metals and soil microbes[J]. Environmental Chemistry Letters, 2017, 15(1): 65-84. doi: 10.1007/s10311-016-0587-x [74] WANG B J, BARAHONA M, BUCK M. A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals[J]. Biosensors and Bioelectronics, 2013, 40(1): 368-376. doi: 10.1016/j.bios.2012.08.011 [75] BEREZA-MALCOLM L T, MANN G, FRANKS A E. Environmental sensing of heavy metals through whole cell microbial biosensors: A synthetic biology approach[J]. ACS Synthetic Biology, 2015, 4(5): 535-546. doi: 10.1021/sb500286r -




 下载:
下载: