-
抗生素是一类具有广谱抗菌性的天然或半人工合成的化学物质,种类繁多,主要包括β-内酰胺类、四环素类、氨基糖苷类、喹诺酮类、大环内酯类等[1 − 2],被广泛应用于多种疾病的预防和治疗[3 − 4]. 然而,抗生素的滥用,一方面会导致环境污染,对人类生命健康和生态环境安全构成严重威胁[5];另一方面,也可能造成“超级细菌”的出现,从而限制抗生素的临床应用[1]. 抗生素消耗量大、处置不当或过量排放容易造成其在水体、土壤等环境介质中的富集,因此,食品和环境中抗生素的快速检测对于维持生态系统稳定和保护人类健康具有重要的意义[6].
传统的抗生素检测方法主要包括液相色谱法[7 − 8]、气相色谱法、液相色谱质谱联用技术[9 − 10]等,都存在仪器昂贵、操作复杂、需要专业技术人员操作等问题,难以满足抗生素快速检测的需求. 虽然酶联免疫分析法[11]、毛细管电泳法[12]等分析技术具有很高的灵敏度和准确度,但仍存在样品前处理步骤复杂、试剂消耗量大、需要专业的技术人员等问题[13]. 因此,开发简便、快速、灵敏的抗生素检测新技术具有重要的现实意义. 近年来,生物传感技术以其集成简单、便携、检测快速且结果易读取等优势,成为检测领域的研究热点[14]. 生物传感器是通过产生可检测的物理或化学信号(如颜色、光信号、电信号等)来实现对不同目标物的定性和定量分析,通常由分子识别元件、信号转导元件、信号输出或放大元件3个部分组成[15],能够将生物化学反应转换为可读取的信号. 传统的生物传感器主要以抗体作为分子识别原件,存在稳定性差、检测目标单一等问题. 适配体与抗体相比,具有易于合成和修饰、热稳定性好、靶标范围广、温度和pH范围宽、无毒性和免疫原性等特点[16 − 17]. 因此,适配体可取代抗体,作为理想的分子识别与传感元件[18 − 19],进而,适配体传感器为抗生素等小分子污染物的检测提供了新思路.
本文综述了氨基糖苷类、四环素类和β-内酰胺类等多种抗生素的适配体筛选,首先总结了特异性结合卡那霉素、四环素、氯霉素等常见抗生素的适配体筛选过程及检测机制,其次概述了近年来用于抗生素检测的适配体传感器研究进展,并重点介绍了基于比色、光学及电化学的新型适配体传感器在抗生素检测中的应用.
-
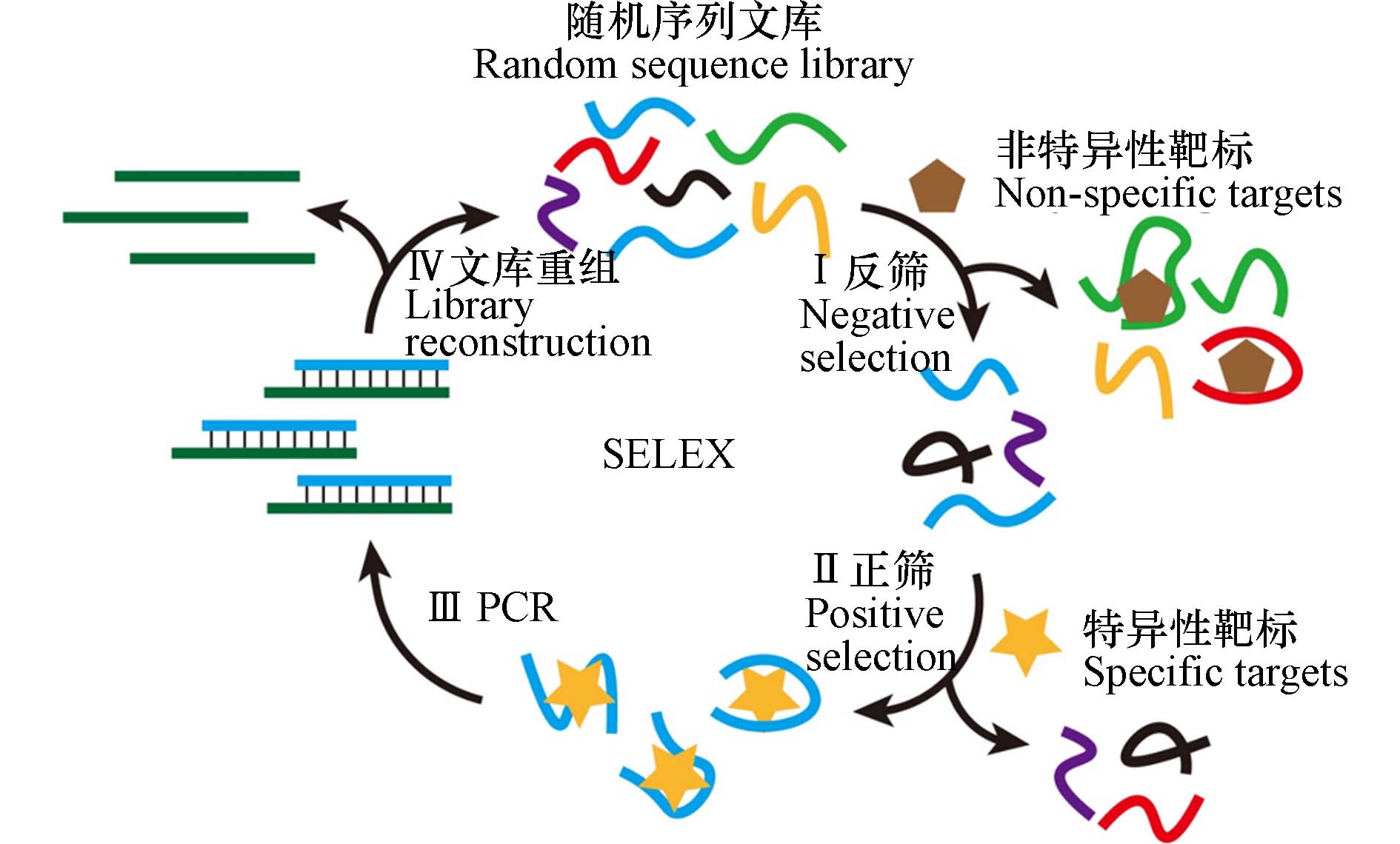
适配体是从随机文库中通过体外筛选技术(SELEX)进化而来,是一种单链寡核苷酸序列,长度通常在20—80个核苷酸之间. 适配体能结合特定的靶标分子(如核酸、蛋白质、金属离子、小分子等)[20 − 21],通常以解离常数(kd)来评估适配体与靶标分子的结合亲和力,kd值越小,代表其亲和力越强[22 − 24]. 适配体的SELEX筛选策略如图1所示,该方法包含体外选择和目标序列扩增等多个循环步骤. 以DNA适配体筛选为例,首先构建包含大量DNA序列(约1014—1016条)的随机化文库,利用非靶标孵育(反筛)的方式,排除非特异性结合的序列,再利用靶标孵育(正筛)筛选能够与靶标结合的目标DNA分子,对其进行洗脱、提取,然后经聚合酶链式反应(PCR)进行目标DNA分子的扩增,获得用于下一轮筛选的次级文库. 通过上述四个步骤的往复循环,实现特异性结合DNA分子的富集.
利用SELEX技术最先获得的适配体是RNA分子,但是RNA易被核酸酶降解,且稳定性较差,限制了其在适配体传感器中的应用. 因此,研究人员将关注点转向稳定性更高的DNA分子,利用筛选技术,得到了一系列能与各种靶标分子特异性结合的DNA适配体. 适配体与靶标分子之间相互作用主要包括氢键、范德华力、疏水相互作用和形状互补等[25 − 26],使得适配体能够与特定的靶标分子结合并发生构象变化[27 − 28],从而形成具有靶标结合核心位点的三维折叠结构,如G-四链体和发夹结构等[29 − 30]. 此外,适配体的空间构型与核酸分子的长度、初始序列排列方式、外界环境等因素密切相关. 适配体是通过茎环结构来实现对抗生素分子的特异性识别,这种结构通常由保守序列组成,作为核心结合位点,具有稳定性高、环境适应性强等特点. 适配体因其独特的优势,在发展适配体传感技术中得到了广泛应用[16 − 17]. 目前,利用SELEX技术,已经筛选出对多种抗生素具有特异性识别功能的适配体,并已建立了各种生物传感策略[2,29,31].
-
氨基糖苷类抗生素是链霉菌来源的一类重要的抗菌药物,其分子结构中包含链胺环,对革兰氏阴性及阳性细菌均具有较好的抗菌活性. 这类抗生素作用于核糖体的30S亚基,导致翻译过程中的错误读取,从而抑制蛋白合成,在医学、畜牧业等领域具有重要的应用价值[32 − 33]. 卡那霉素(KANA)、链霉素(STR)、妥布霉素(TOB)、庆大霉素(GEN)等都是比较常见的氨基糖苷类药物[34].
Wang等[35]最早在1995年成功分离结合妥布霉素(TOB)的RNA适配体(X1和J6),并鉴别出截短的适配体(X1SL和J6SL). 该适配体能够结合TOB、KANA和GEN,kd值均在2—12 nmol·L−1,选择性不佳. 随后,Morse等[36]成功选择对TOB具有良好特异性的RNA适配体(BA14-1和BA14-2). 相较于DNA而言,RNA具有稳定性差的固有性质,普适性较低. Spiga等[37]提出了一种新的基于DNA的捕获-SELEX策略,所选择最佳适配体的kd为200 nmol·L−1,该方法在无标签SPR实验中可检测到TOB的浓度比临床低20倍,表明该适配体适用于小型药物分子检测.
针对卡那霉素(KANA)适配体体外筛选的研究,Song等[38]采用KANA固定化的琼脂糖珠亲和层析法进行体外筛选,获得了DNA适配体(Ky2),对KANA、KANA-B和TOB均具有较高亲和力,kd值分别为78.8 nmol·L−1、84.5 nmol·L−1和103 nmol·L−1,Ky2已被应用于不同类型的生物传感器[39]. Stoltenburg等[40]利用捕获-SELEX将DNA固定于磁珠上,获得了KANA响应的DNA适配体(3-19),但3-19包含97个碱基,比先前的适配体长得多,不具备合成优势以及生物传感应用潜力. 此外,Sanford等[41]报道了一种结构转换适配体筛选方法,利用限制性内切酶,分离经过靶诱导的短互补链位移的适配体. 经过筛选获得了KANA-A的适配体(K16-1),进一步序列截短优化,获得了包含21个碱基的K16-1c,能够用于结构转变型生物传感器. 该传感器对KANA-A的检测范围为90 μmol·L−1—10 mmol·L−1,并且相比其他3种氨基糖苷类抗生素,具有较高的选择性. 此外,针对其他氨基糖苷类抗生素的适配体筛选也报道了诸多研究,如链霉素(STR)[42 − 44]、新霉素(NEO)[45 − 47]、庆大霉素(GEN)[48]等,表1列出了所有氨基糖苷类抗生素适配体的名称、类型、序列信息及解离常数.
-
四环素类抗生素(TCs)是一种链霉菌素,能够抑制细菌的蛋白质合成,具有抗菌活性. 因其具有治疗效果优良、价格低廉等特点,被广泛用于人类或动物感染的治疗[51 − 52]. 常见的四环素类抗生素有四环素(TET)、土霉素(OTC)、金霉素(CTC)、强力霉素(DOX)等[53 − 54].
对四环素(TCs)响应型适配体的研究也始于RNA分子,Berens等[55]通过体外筛选分离出识别TET的RNA适配体(cb28),研究了TET与cb28之间的相互作用模式,证明了两者结合后适配体的构象变化. 研究发现cb28不识别OTC、CTC等其他四环素类抗生素,能够与TET特异性结合,选择性良好,但是结合亲和力在微摩尔浓度水平. Xiao等[56]通过体外筛选获得了亲和力更高的TET响应型RNA适配体,TET以镁离子螯合物的形式与适配体结合,其kd值为0.8 nmol·L−1. 由于RNA的不稳定性,易被环境中的核酸酶降解,在生物传感应用中比较受限,为提高用于抗生素检测的适配体适用性,识别四环素的DNA适配体受到广泛研究.
Niazi等[57]利用体外筛选技术,筛选出了对土霉素(OTC)具有高亲和力和特异性的单链DNA适配体,编号为No.4、No.5和No.20的3个适配体具有较强的亲和力,kd分别为9.61 nmol·L−1、12.08 nmol·L−1和56.84 nmol·L−1,对OTC的选择性为72%—76%. 随后,通过修饰-SELEX意在获得TET特异性的DNA适配体[58],作者分别用OTC、TET和DOX涂层的磁珠作为正靶标和反靶标,最终获得了7条亲和力较高的适配体,它们的kd值为63—483 nmol·L−1,对TCs的亲和力大小为:OTC﹥TET﹥DOX. 随后,Kwon等[59]通过删减适配体与靶标结合时的冗余序列,最终获得8个核苷酸长度的DNA适配体(A2),A2能够识别4种不同类型的TCs,kd值为1.067 nmol·L−1,表明序列截短能够提高适配体的结合亲和力. 通过对筛选策略的优化,Zhao等[60]利用DNA文库固定化捕获游离的靶标,筛选获得了OTC特异性的无标记DNA适配体(OTC5),其对OTC、TET、DOX具有类似的结合亲和力和通用性.
用于TCs检测的RNA适配体与DNA适配体相比[61],RNA适配体具有更低的kd,Kwon等[59]通过对DNA适配体序列的截短优化,成功获得了亲和力相近于RNA的DNA适配体. 通过以上综述,发现多数TCs的DNA适配体均与OTC具有较强的结合作用,仍未发现对TET具有良好选择性的DNA适配体,虽然这些适配体具有TCs检测的通用性,但在区分TCs类似物时仍存在局限. 表2列出了所有四环素类抗生素适配体的名称、类型、序列信息及解离常数.
-
β-内酰胺类抗生素因其高效、低毒性、可生物降解等优点而被广泛应用于临床治疗[66],其作用机制是通过阻断细胞壁多肽间的交联,特异性地靶向具有黏液层的原核生物. 该类抗生素以β-内酰胺环为特征,常见的有氨苄青霉素(AMP)、青霉素(PEN)等[67].
Song等[68]报道了氨苄青霉素(AMP)特异性适配体,并建立了金纳米颗粒双荧光比色传感平台,利用磁珠SELEX技术,获得了AMP7、AMP17和AMP18等3种DNA适配体,其kd分别为9.4 nmol·L−1、13.4 nmol·L−1和9.8 nmol·L−1. 此外,Paniel等[69]使用捕获-SELEX技术获得了对青霉素G(PEN-G)具有选择性的适配体,该SELEX技术将DNA文库固定于载体上,捕获游离于溶液中的靶标,结果表明,该适配体能够与AMP、阿莫西林等同类抗生素结合,但结合亲和力较弱. 随后,Lee等[70]通过还原GO-SELEX(rGO-SELEX)鉴定了一种高亲和力的PEN特异性适配体,kd值为383.4 nmol·L−1. Wang等[71]采用磁珠SELEX法筛选出4个头孢喹肟(CFQ)特异性DNA适配体,鉴定出适配体W1,kd值低至40.13 nmol·L−1,并建立了高灵敏度检测牛奶样品中CFQ残留的荧光适配体传感器. 表3列出了所有β-内酰胺类抗生素适配体的名称、类型、序列信息及解离常数.
-
适配体传感器有多种信号读取模式,如光学、电化学、质量变化、热量变化等,其中基于比色、光学和电化学的适配体传感器得到了广泛的研究与应用. 本节着重讨论了比色、光学和电化学适配体传感器的基本原理,并对其在抗生素测定方面的应用进行了阐述.
-
在不同的传感方法中,比色适配体传感器是最简单的一种,该方法是通过与已知物质浓度的色标相对比,以测定液相中的目标物浓度. 比色传感具有反馈快速、成本效益、可视觉检测以及无需特殊仪器等优势,在生物传感领域受到越来越广泛的关注.
金属纳米颗粒由于其独特的尺寸和间距,具有特殊的光学特性,基于金纳米颗粒(AuNPs)和银纳米颗粒(AgNPs)的比色适配体传感器被广泛研究[72]. AuNPs具有合成简单、消光系数高等特点,提高了比色分析的灵敏度[73]. 适配体负载于AuNPs表面时,阻止了AuNPs团聚[74],此状态下的粒子直径一般小于40 nm,AuNPs呈现红色. 当靶标分子存在时,适配体发生构象变化,由随机形状的线性结构转变为折叠的刚性结构,使得AuNPs表面的适配体发生脱附,导致AuNPs聚集,其颜色随着粒子直径的增加而变为蓝色或紫色. AuNPs聚集过程中呈现的颜色变化被广泛应用于目标分析物的比色检测[75].
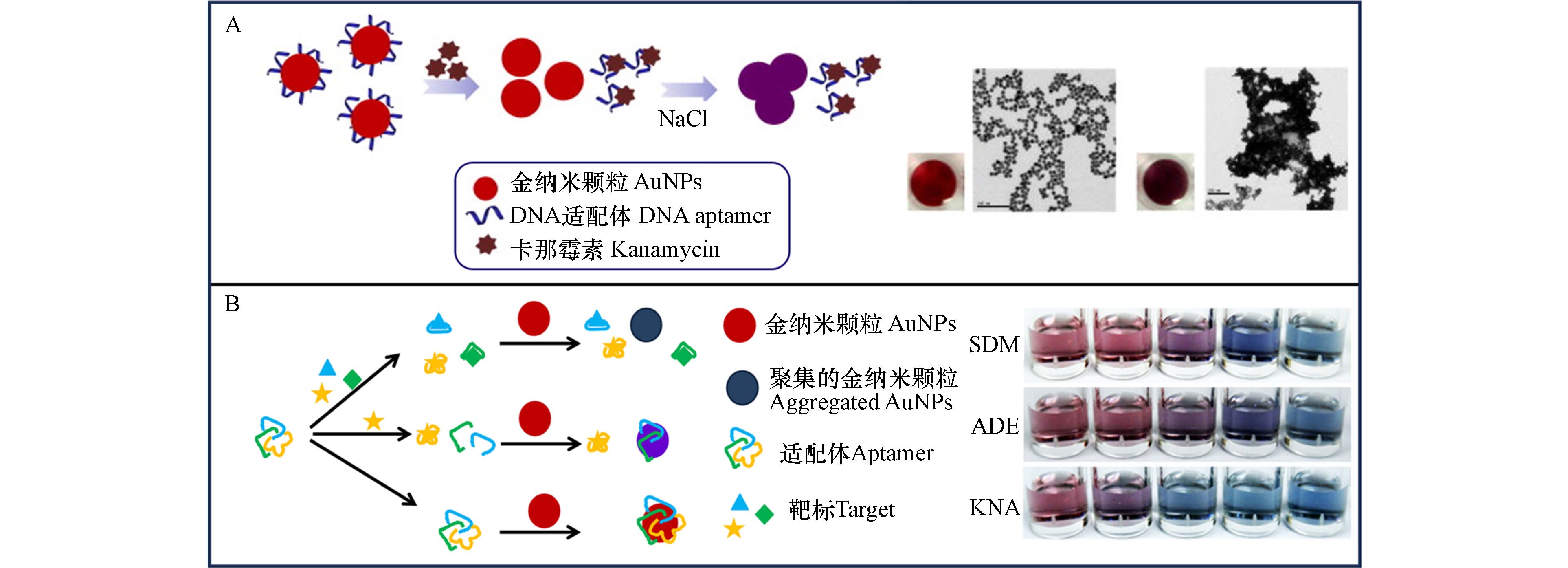
Song等[38]将卡那霉素适配体固定于AuNPs表面,构成基于比色的适配体传感器. 当卡那霉素存在时,盐离子的加入使得AuNPs聚集,导致颜色由红色变为紫色,如图2A所示. 该比色方法成本低、操作简单,并且可以通过肉眼观察颜色变化,被广泛用于卡那霉素检测[38,76]. 此外,将3种不同的DNA适配体和AuNPs组合,构建能够同步检测磺胺二甲氧嘧啶(SDM)、卡那霉素(KNA)和腺苷(ADE)的适配体传感器[77],如图2B所示. 与单目标检测相比,该方法的灵敏度未发生改变,ADE、SDM和KNA的检出限分别为500 ng·mL−1、500 ng·mL−1和100 ng·mL−1.
除基于AuNPs的显色方法外,Wang等[78]开发了一种基于间接竞争酶联(ic-ELAA)的适配体传感方法,用于检测食品中的四环素残留. 如图3A所示,该方法利用已经获得的DNA适配体[62],对蜂蜜中的四环素进行识别和检测. 随后,该课题组提供了一种改进的直接竞争酶联(dc-ELAA)适配体测定方法,对蜂蜜中四环素检测的线性检出范围为0.1—
1000 ng·mL−1,检出限为0.0978 ng·mL−1. 与ic-ELAA的适配体传感器相比,dc-ELAA具有检测限好、操作简单等优点,可用于食品中四环素的高通量检测[64]. 随后,Wang等[79]设计了一种基于适配体的液晶检测卡那霉素的方法,如图3B所示,当卡那霉素存在时,与适配体相互作用形成G-四链体结构,破坏了表面液晶的定向排列,使得薄膜上的颜色由粉红色变为绿色,可用正交偏光镜视觉观察检测结果,特异性高,检出限为1 nmol·L−1.此外,G-四链体是一种具有过氧化物酶性质的脱氧核酶(G4 DNAzyme),与血红素(hemin)结合后能够进行催化反应,使得2-2’-偶氮-双(3-乙基苯并噻唑啉)-6-磺酸(ABTS)或3,3’,5,5’-四甲基联苯胺(TMB)发生颜色变化,该方法是一种备受关注显色策略. Cui等[80]报道了一种基于靶激活的G4 DNAzyme偶联双NESA(双剪切酶信号放大)的检测方法(图3C),靶标与适配体结合后产两个富G的分裂片段,进而诱导双NESA反应,同时G4 DNAzyme作为生物催化剂,催化H2O2介导的氧化反应,使ABTS2-转化为ABTS-,从而发生颜色变化,因此可以通过肉眼进行视觉观察.
-
光学适配体传感器以适配体为分子探针,通过多种光学分析方法,将采集到的信号转化为可检测的光学信号. 光学传感技术因其响应快速、灵敏度高、操作简单等特点,在抗生素分析中得到广泛的应用. 目前,光学适配体传感器主要有荧光、化学发光、表面增强拉曼散射(SERS)、表面等离子体共振(SPR)等五大类,本文主要介绍基于荧光、化学发光和拉曼散射的光学适配体传感器.
-
荧光是一种常见的光学检测方法,由于其灵敏度高、响应快速、操作简便等特点,被广泛用于适配体传感领域. 由于适配体本身不具备荧光特性,故通常采用荧光基团或猝灭基团标记的适配体作为荧光探针. 纳米粒子因其独特的优势,可用作荧光适配体传感器的改性薄膜,有利于提高传感器的检测性能. 利用适配体与靶标结合后的构象变化,发生荧光共振能量转移(FRET)[81]从而使得荧光信号产生增强或猝灭,胶体金纳米颗粒(AuNPs)[82]上转换纳米颗粒(UCNPs)[83]等已被用于构建荧光适配体传感器.
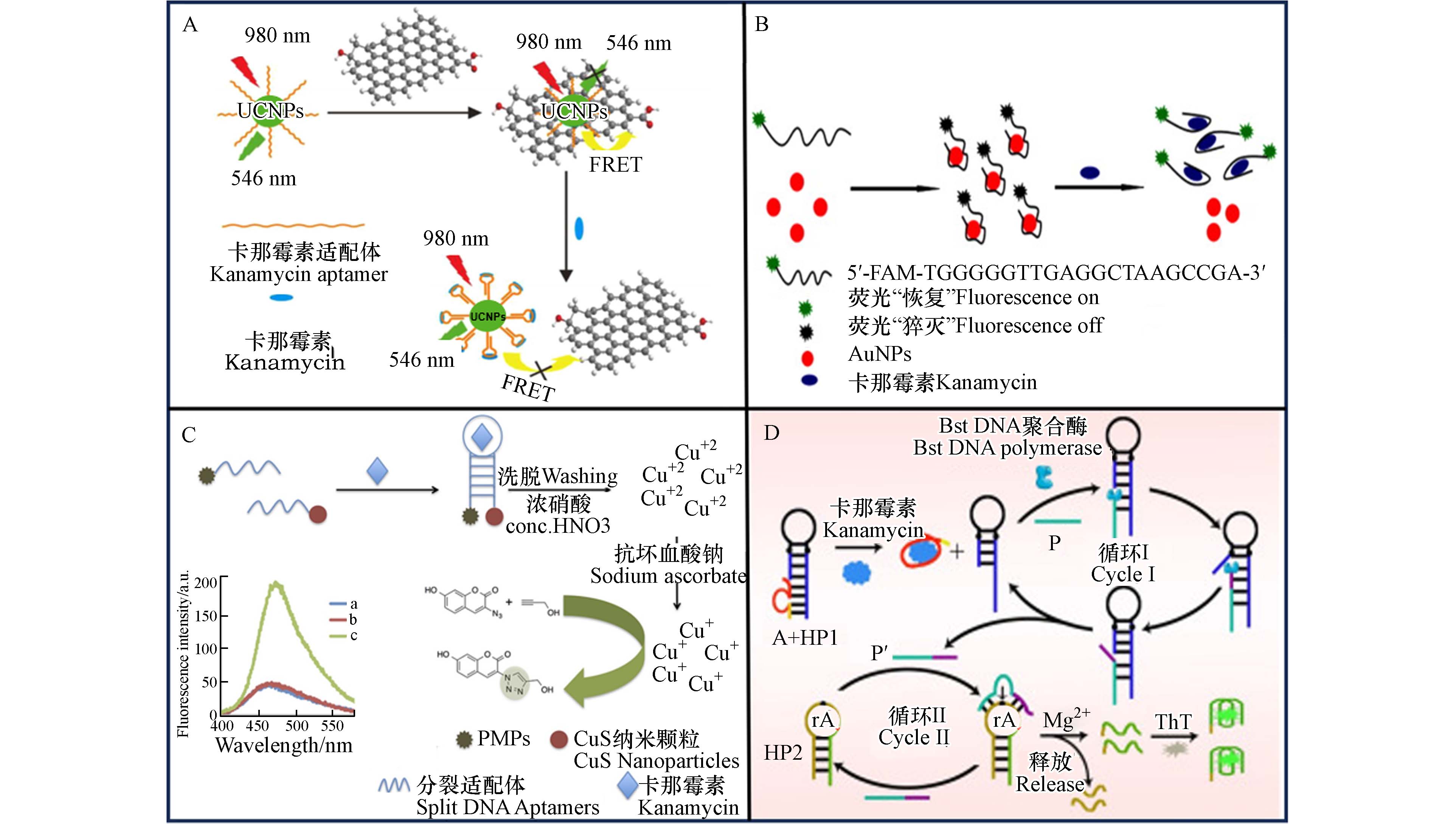
Li等[82]报道了一种用于卡那霉素检测的超灵敏FRET适配体传感器. 如图4A所示,该研究通过适配体与石墨烯之间的π-π堆叠相互作用,拉近UCNPs与石墨烯之间的距离,从而启动FRET过程,导致UCNPs荧光猝灭. 向UCNPs-适配体-石墨烯配合物中加入卡那霉素,适配体构象转变成发夹结构,能量转移被阻断,使得荧光恢复. 以AuNPs为基底,在其表面固载适配体分子,从而被用作分子传感标签,构建光学适配体传感器[75]. Chen等[84]提出了一种基于FRET的适配体传感器用于检测卡那霉素A,如图4B所示,该传感器采用AuNPs作为DNA纳米载体和荧光猝灭剂,对卡那霉素的检测限为0.3 nmol·L−1. Belal等[85]开发了一种荧光响应的卡那霉素检测方法,如图4C所示. 当卡那霉素的存在时,适配体形成发夹结构,释放出的Cu2+经抗坏血酸钠还原为Cu+,催化点击化学反应,从而生成荧光物质(1,4-双取代-1,2,3-三氮唑),在0.04—20 nmol·L−1范围内,荧光信号与卡那霉素浓度的对数值呈良好的线性关系,该方法重现性好且选择性高.
一般来说,适配体上荧光基团的标记会降低其与靶标分子之间的亲和力[87],无标记适配体传感器的研究越来越广泛,另外,信号放大策略可以有效提高适配体传感器的检测灵敏度. Zhou等[86]建立了一种用于牛奶中卡那霉素检测的荧光传感平台,如图4D所示. 适配体与卡那霉素结合后,启动PER,在Bst-DNA聚合酶辅助下,自发合成Mg2+依赖型的DNAzyme,循环切割信号发夹底物中的RNA位点,从而释放大量游离的G-四链体片段. 有机染料硫黄素T (ThT)与之结合后,产生明显增强的荧光信号. 该传感器对卡那霉素的检出限为0.36 nmol·L−1.
-
化学发光法是化学反应过程中物质吸收化学能而产生的光辐射,其共振能量转移来源于化学发光底物的氧化. 与荧光法相比,化学发光法不需要外界的激发源,是一种性能良好的分析方法,具有灵敏度高、设备成本低、检测线性范围宽等优势.
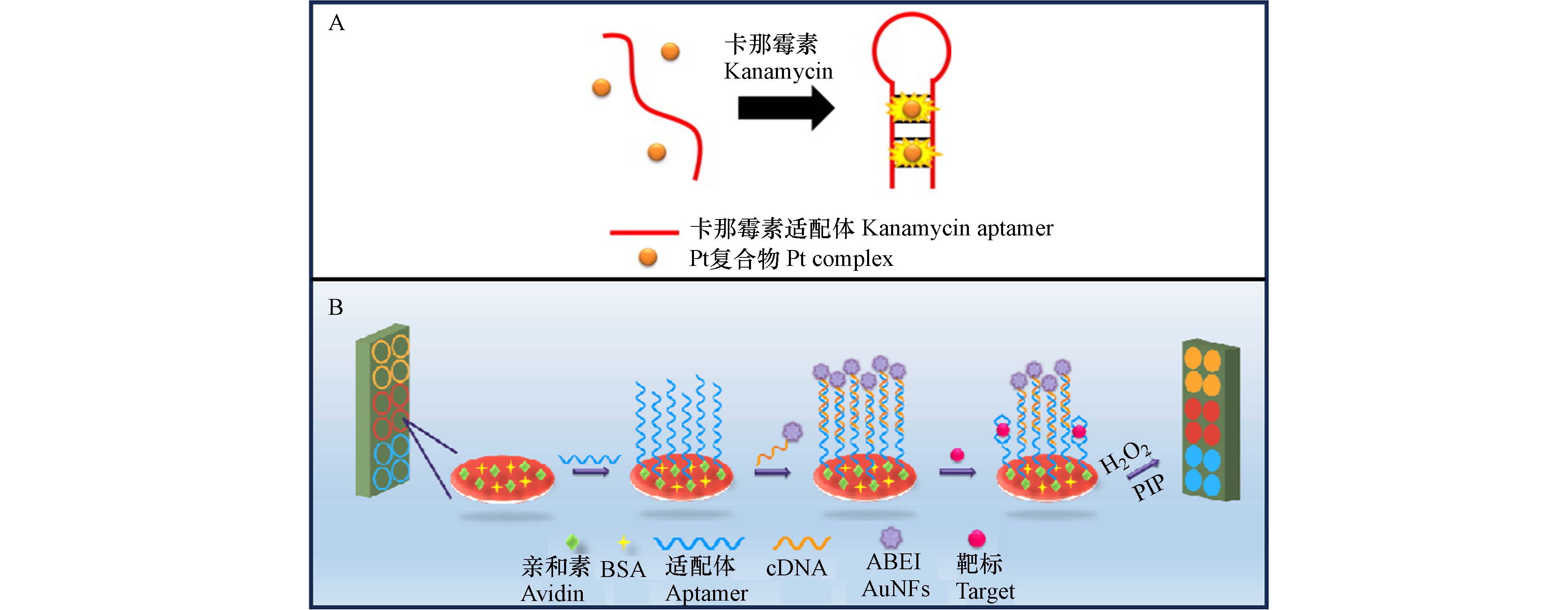
Leung等[88]报道了一种检测水体中卡那霉素的化学发光方法. 该方法利用方-平面发光铂(Ⅱ)配合物和卡那霉素适配体,如图5A所示,当卡那霉素不存在时,适配体的随机三维结构与铂(Ⅱ)配合物表现出较弱的相互作用,从而产生较低强度的发射信号. 当加入卡那霉素后,适配体的构象转变为双螺旋发夹结构,利于发光铂(Ⅱ)配合物嵌入适配体,从而实现卡那霉素的发光响应信号增强,在545 nm处检测出发光响应,检出限为143 nmol·L−1. Hao等[89]建立了一种同时检测3种抗生素的高灵敏、高特异性的多重化学发光法,如图5B所示. 硫代杂交互补链(cDNA)修饰的N-(4-氨基丁基)-N-乙基异米诺尔(ABEI)功能化金纳米化结构作为信号探针,将土霉素、四环素、卡那霉素适配体固定在酶标板底部作为分子识别探针. 靶标和信号探针可以竞争性地与固定化适配体结合,因此,基于ABEI-H2O2-P-碘苯酚(PIP)的稳态化学发光系统可以同时检测土霉素、四环素和卡那霉素. 该方法的化学发光强度分别与3种靶标浓度的负对数相关,三者的检出范围分别为0.05—5 ng·mL−1、0.05—5 ng·mL−1、0.05—0.5 ng·mL−1,检出限分别为0.02 ng·mL−1、0.02 ng·mL−1、0.002 ng·mL−1. 采用加标法对牛奶中抗生素成分的测定能力进行验证,结果与商业ELISA方法结果一致.
-
表面增强拉曼散射(SERS)是一种振动光谱技术,能够识别分子的化学结构、官能团等信息,已经成为一种有效的、非侵入性的、现场检测的生物分析方法[90]. 基于SERS的适配体传感器已广泛应用于重金属、毒素、农药、抗生素等多种分析物的检测[91 − 92].
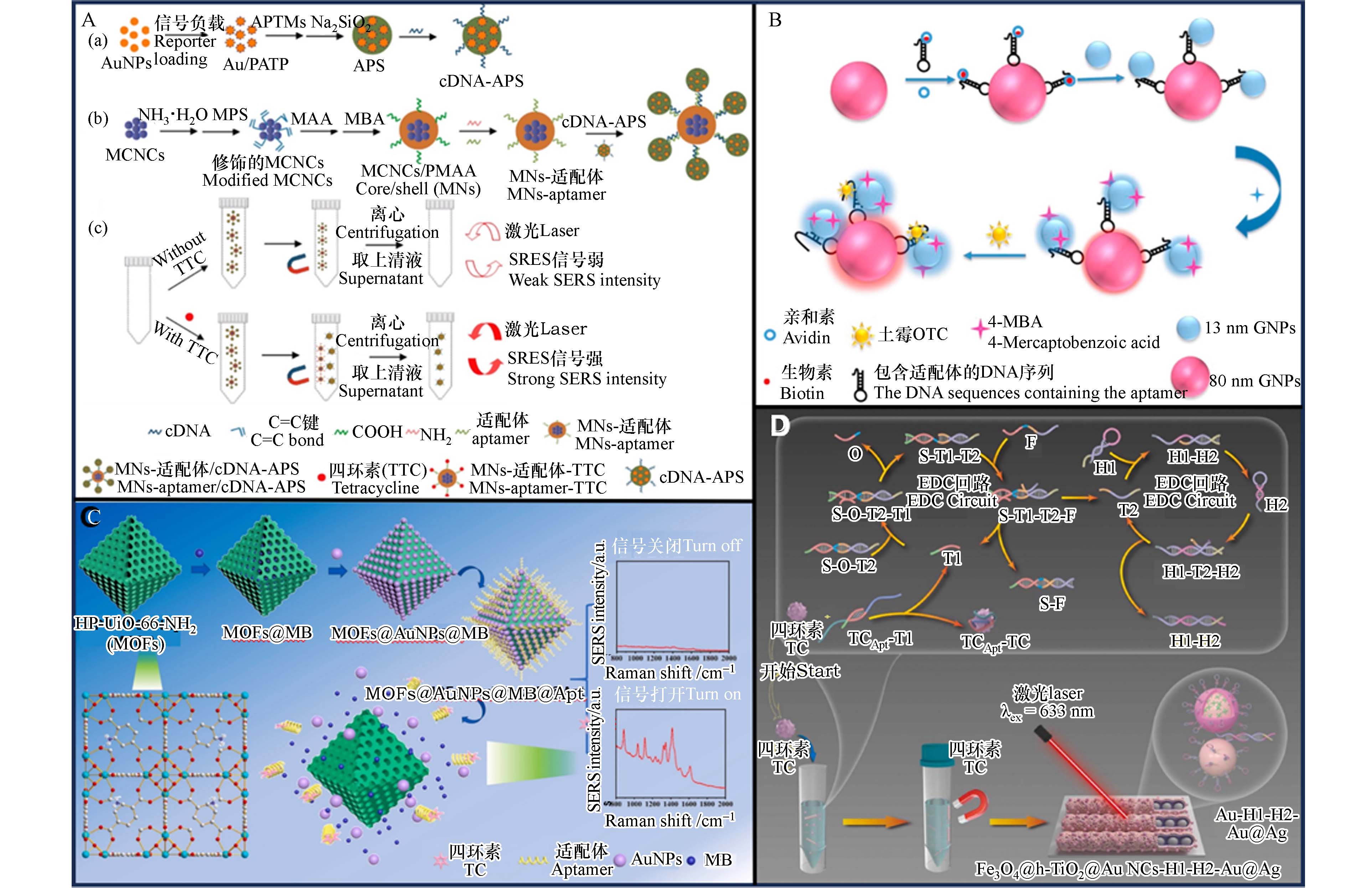
Li等[92]设计了一种基于SERS的适配体传感器,用于检测四环素. 如图6A所示,该传感器靶向磁性纳米球,以适配体-共轭磁铁矿胶体纳米晶簇(MCNCs)-聚甲基丙烯酸(PMAA)磁性纳米球(MNs)作为识别元件,以Au/PATP/SiO2(APS)作为标记分子. 当加入四环素分子后,适配体与之结合,cDNA-APS被释放到溶液中,在磁力作用下纳米颗粒聚集,与信号分子分离,上清液进而显示出增强的SERS信号. 该检测方法的检测范围较宽,线性检测范围为2.25×10−6—2.25×10−1μmol·L−1,且具有良好选择性. 同时,Meng等[93]提出了另一种基于SERS的土霉素检测方法,检测原理如图6B所示,该方法是基于偶联有DNA序列的AuNPs(直径分别为13 nm和80 nm)之间的拉曼热点而构建的. 将拉曼信号分子(4-MBA)修饰于13 nm的AuNPs表面,OTC暴露后,适配体序列优先与OTC结合,并与其互补片段解旋,从而拉近13 nm的AuNPs与80 nm AuNPs之间的距离,这种相互作用产生的热点越强,SERS信号越高. SERS信号与OTC浓度呈正相关,检测范围为9.27×10-11—9.27×10−7μmol·L−1,检出限低至8.77×10-12 μmol·L−1.
随后,Li等[94]采用适配体门控HP-UUO-66-NH2纳米通道策略,开发了一种用于四环素灵敏检测的目标响应释放式SERS适配体传感器,如图6C所示. 作者将亚甲基蓝(MB)和AuNPs分别作为信号探针和封盖剂加载到合成的金属有机骨架(HP-UiO-66-NH2MOF)中. 当四环素存在时,适配体的“分子门”被打开,从而导致MB和AuNPs的“货物释放”. 因此,可以通过监测上清液SERS强度的变化来确定TC的浓度. 此外,该方法在实际样品(牛奶和猪肉)加标实验中具有良好加标回收率(93.23%—108.79%),证明该方法在抗生素检测中具有巨大潜力. Lv等[95]基于适配体、无酶DNA回路和SERS技术,构建了用于四环素(TC)灵敏检测的级联扩增SERS适配体传感器(图6D). 将DNA发夹H1和H2分别结合到制备的Fe3O4@hollow-TiO2/Au纳米链(Fe3O4@h-TiO2/Au NCs)和Au@4-MBA@Ag纳米粒子上,构建捕获探针和信号探针. 在最佳条件下,该适配体传感器对TC有明显的线性响应,最低检出限为15.91 pg·mL−1. 此外,作者所提出的级联扩增传感策略具有良好的特异性和存储稳定性,通过实际样品中TC的检测验证了其实用性和可靠性.
-
电化学适配体传感器的核心在于固定在电极表面的氧化还原探针标记的适配体,当靶标存在时,适配体的构象变化会改变探针与电极表面的距离,从而发生或阻断电子传递,实现电化学读数[96],使得所读信号与分析物浓度呈正相关或负相关的信号模式[97]. 电化学适配体传感器具有无标签、简单、实用、灵敏等特点,在抗生素检测中的得到了广泛关注. 电化学传感器根据其原理大致分为基于阻抗、电流和电势等3种类型,下文介绍了基于阻抗和电流的电化学适配体传感器及代表性研究.
-
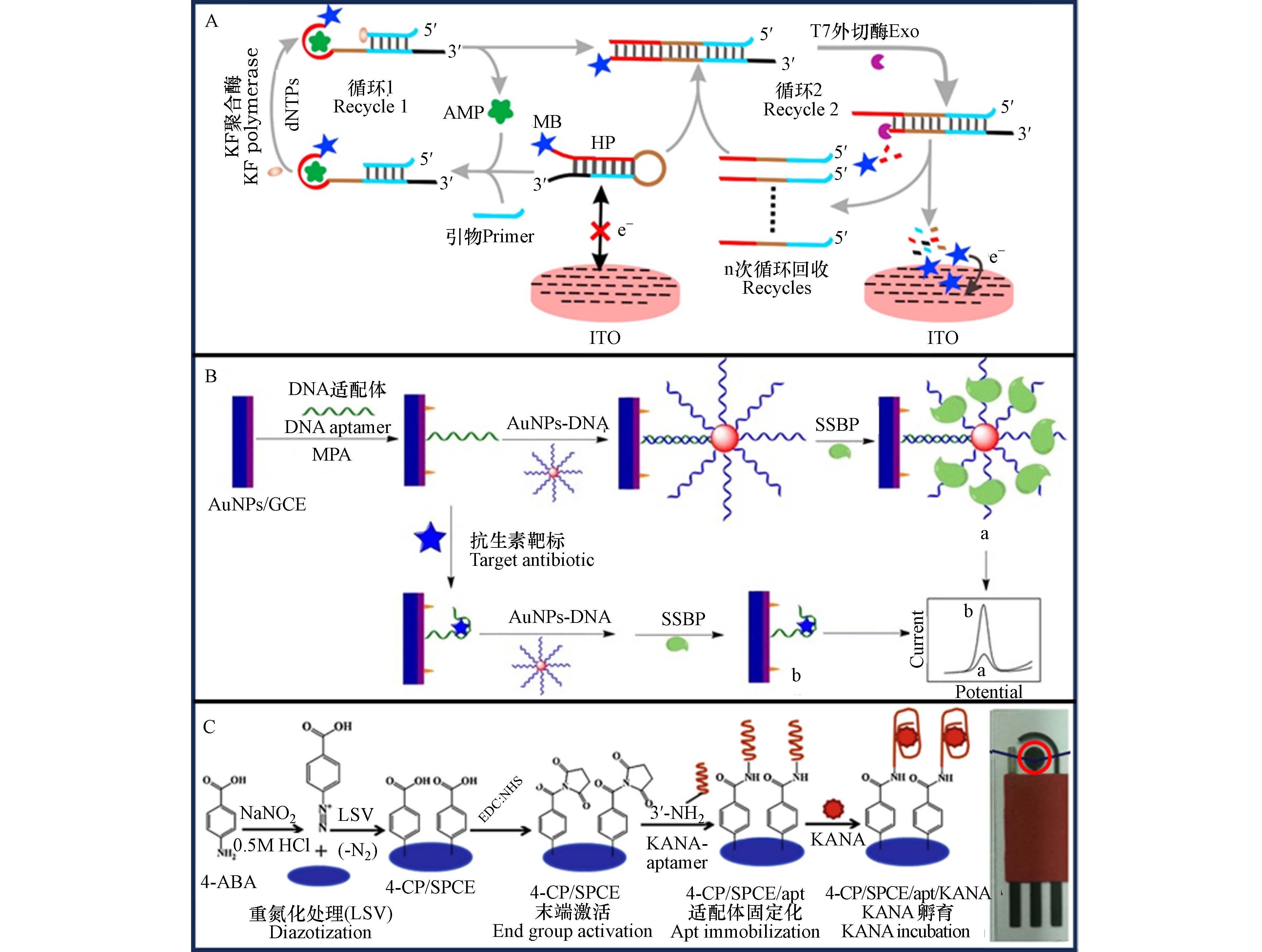
基于阻抗的电化学适配体传感器工作原理是感应适配体与靶标相互作用时溶液中的阻抗变化. Dapra等[98]和Rosati等[99]设计了阻抗生物传感器,应用于氨苄青霉素(AMP)和卡那霉素(KANA)检测. 为了提高适配体传感器对AMP的响应速度和信号放大,Wang等[100]利用聚合酶和切口酶设计了用于抗生素残留检测的电化学适配体传感器,如图7A所示,该传感器基于靶向诱导和T7外切酶辅助的双循环信号放大策略,依赖于发夹探针和单核苷酸对带负电荷的氧化铟锡电极的扩散率差异,该方法对AMP的检出限低至4 pmol·L−1. 在现有的电化学传感器分析中,基于DNA功能化AuNPs(DNA-AuNPs)和ssDNA结合蛋白(ssDNA-BP)的适配体传感器具有更高的灵敏度. 该传感器[101]的检测原理如图7B,当适配体与AuNPs上的靶DNA杂交时,ssDNA-BP通过与ss-DNA的特异性相互作用被捕获在电极表面,导致氧化还原探针Fe(CN)63-的电化学信号下降. 当AMP存在时,适配体与AMP的结合阻断了DNA-AuNPs和ssDNA-BP的偶联,其对AMP的检出限低至0.38 pmol·L−1.
Sharma等[102]设计了一种用于卡那霉素检测的便携式阻抗适配体传感器. 如图7C所示,该传感器将氨基功能化的卡那霉素适配体通过NH2-COOH相互作用固定在丝网印刷碳电极(SPCE)表面,适配体与卡那霉素的相互作用抑制了法拉第反应,从而增加了电子转移阻力. 为进一步提高电化学适配体传感器的检测特异性和灵敏度,已经报道了许多优秀的研究[103].
-
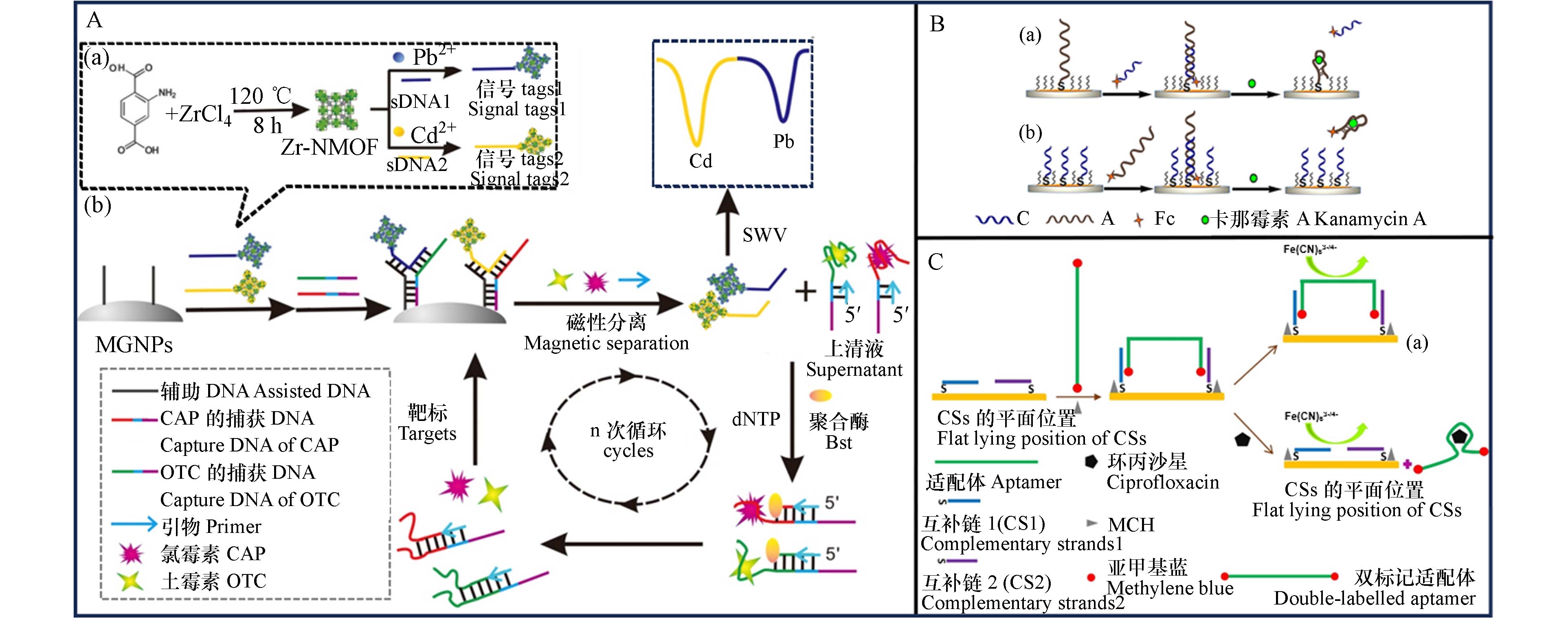
基于电流的电化学适配体传感器工作原理是感应适配体发生配对时氧化还原电流的变化. Chen等[104]利用基于电流的电化学传感器,如图8A所示,当靶标存在时,捕获DNA的构象变化使Y-DNA探针解体,从而信号标签释放到上清液中,实现了多种抗生素的检测,同时,捕获DNA的裸露序列在聚合酶的作用下,触发靶循环诱导聚合的启动子,从而实现信号放大,对氯霉素和土霉素的检出限分别为33 fmol·L−1和48 fmol·L−1.
大多数目标诱导链位移分析的信号转导依赖于适配体的构象变化,尽管检出限很低,但限制了生物传感器的应用. Liu等[105]报道了一种信号探针位移电化学适配体传感器(SD-EAB),如图8B所示,这种信号转导方式仅由适配体及其靶标之间的亲和结合诱导,完全依靠适配体的构象变化,典型的SD-EAB由固定在硫代捕获探针(短的互补链或适配体)之间的DNA双链金电极组成,在靶标存在的情况下,信号探针发生位移,并从电极表面释放,导致电流与靶标浓度的对数值呈正比减弱. SD-EAB实现了卡那霉素A的无试剂检测,动态范围为1 nmol·L−1—10 mmol·L−1. 针对其他抗生素药物,Taghdisi Heidarian等[106]建立了检测喹诺酮类药物(FQs)的电化学传感策略,如图8C所示,该方法由作为检测元件的双标记适配体、作为氧化还原剂的亚甲基蓝(MB)以及互补适配体链(CSs)组成. 该策略对环丙沙星的动态检测范围为300—450 nmol·L−1,检出限为100 pmol·L−1,实现了抗生素药物的简单灵敏检测.
-
电化学发光(ECL),也称为电致化学发光,是指在晶片界面处,荧光物质在晶片表面发生能量转移,由基态向激发态转变,并产生荧光信号的过程[107 − 108]. 电化学发光适配体传感器集电化学发光和生物传感器于一体,利用电致化学发光信号,实现对目标物的定量检测,该方法具有操作简单、灵敏度高、响应速度快、本底信号低等优势,在生物分析、环境监测等领域具有广阔的应用前景[109 − 110].
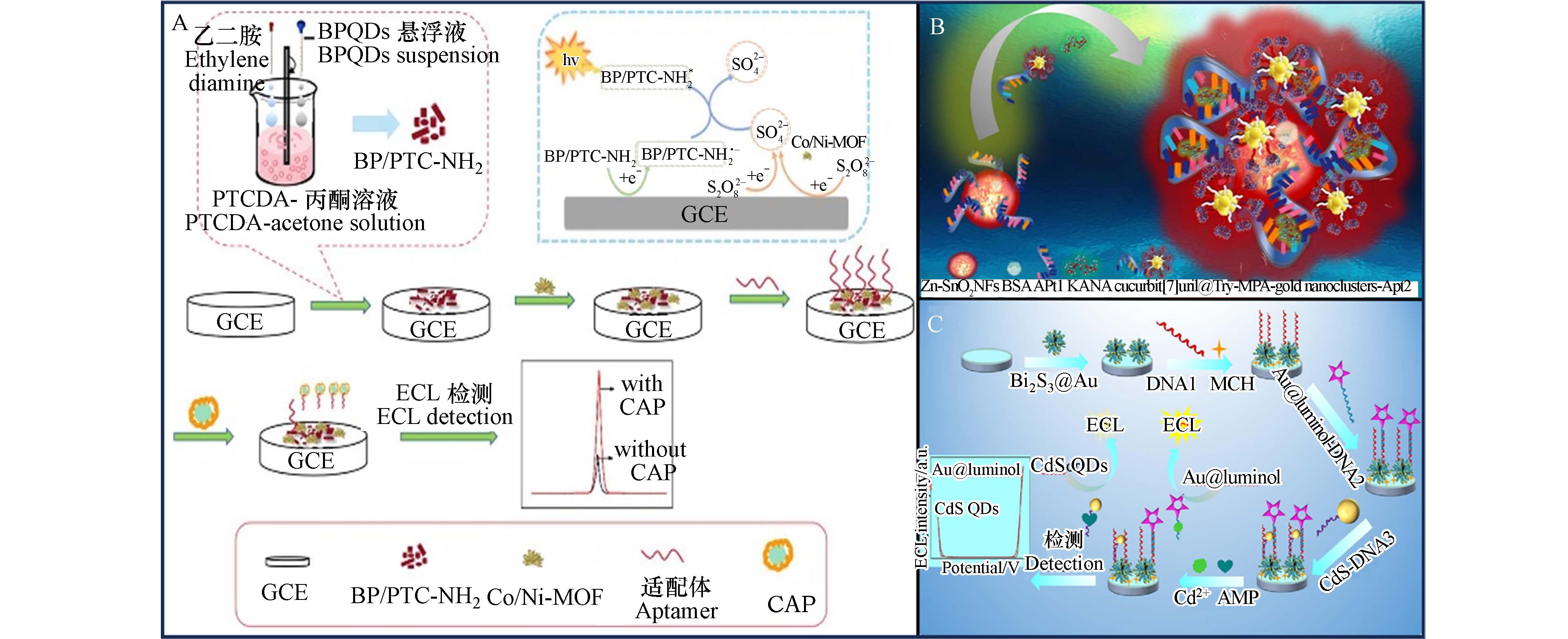
Wen等[111]利用金属-有机框架(Co-Ni/MOF)在苝衍生物(BP/PTC-NH2)和过硫酸氢盐(S2O82-)体系中的增强机制,开发了一种用于检测氯霉素(CAP)的高灵敏度和高选择性的电化学发光适配体传感器. 如图9A所示,作者合成了黑磷量子点(BPQDs),并将其加入PTC-NH2前驱体溶液中,进一步合成了掺杂PTC-NH2的纳米粒子(BP/PTC-NH2),BP/PTC-NH2作为ECL发射体. 适配体识别并结合CAP后,远离传感界面,导致信号增强. 因此,基于这种增强效应和适配体的特异性识别功能,该ECL适配体传感器可测定CAP浓度范围为0.1 pmol·L−1—1.0 μmol·L−1,最低检出限为29 fmol·L−1. 由于复杂的设计和制造工艺妨碍了共价修饰的电化学发光(ECL)发光团能级的改变或能量产生以及电子转移过程,难以提高ECL的检测性能. Zhang等[112]利用色氨酸(Try)和巯基丙酸(MPA)为配体的金纳米团簇(Try-MPA-金纳米团簇),采用非共价键自组装的方法提高了ECL的检测性能,如图9B所示. 以葫芦脲[7](cucurbit[7]uril)处理的Try-MPA-gold纳米团簇作为信号探针,以具有高电子迁移率的掺杂锌SnO2纳米花(Zn-SnO2 NFs)作为电极修饰材料,利用分裂适配体作为捕获探针,建立了用于卡那霉素(KANA)检测的ECL传感器. 改进的分裂适配体传感器对复杂食品底物中的KANA具有良好的灵敏度分析,回收率为96.2%—106.0%.
Zhai等[113]以Bi2S3@Au纳米花为基础纳米材料,分别以Au@luminol(鲁米诺)和CdS量子点作为独立的ECL发射信号,构建了电化学发光(ECL)生物传感器. 如图9C所示,Bi2S3@Au纳米花作为工作电极的衬底,提高了电极的有效面积,加快了金纳米粒子与适配体之间的电子传递速率,为发光材料的负载提供了良好的界面环境. 然后,将Au@luminol功能化DNA2探针作为独立的ECL信号源,在正电位下识别Cd2+,同时将CdS量子点功能化,DNA3探针作为负电位下的独立ECL信号源,可识别氨苄西林(AMP),该设计实现了不同浓度的Cd2+和AMP的同步检测.
-
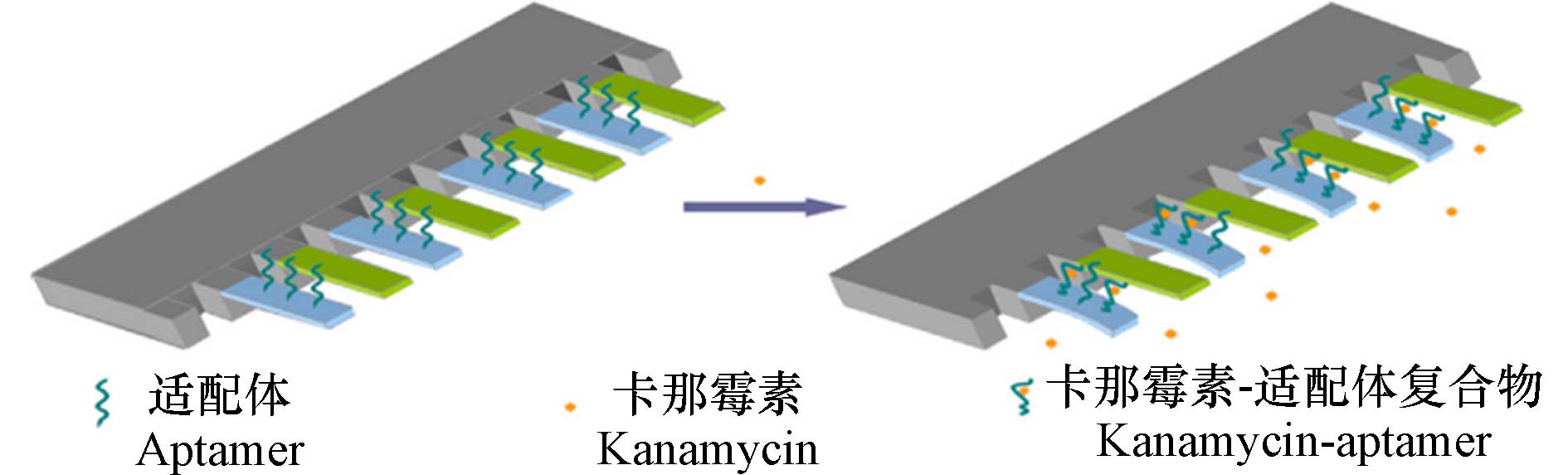
Bai等[114]建立了一种用于卡那霉素检测的无标记适配体悬臂阵列传感器. 悬臂阵列由传感悬臂和参考悬臂组成,如图10所示,这种设计能够直接检测单个悬臂挠度,进而确定传感/参考悬臂的差动挠度. 卡那霉素适配体作为受体分子,使用6-巯基1-己醇(MCH)对感知悬臂梁进行功能化修饰,消除环境干扰. 卡那霉素与适配体之间的相互作用诱导了悬臂表面应力的变化,导致感知悬臂与参考悬臂之间的差异偏转. 在100 μmol·L−1—10 mmol·L−1的浓度范围内,表面应力变化与卡那霉素浓度呈良好的线性关系,相关系数为0.995. 在信噪比为3的条件下,检出限为50 μmol·L−1. 该传感器对新霉素、核糖霉素和氯霉素等抗生素同样表现出良好的选择性.
-
为实现环境中抗生素污染的实时监测,亟需开发一种简便、灵敏、可靠的分析检测技术. 综上所述,适配体传感器可满足此需求,通过多种信号传导机制构建生物传感检测新方法,将其应用于环境、食品、药品等介质中的抗生素污染检测. DNA适配体因其不易被天然的核酸酶降解而具有更强的普适性. 抗生素与相应适配体之间存在相互作用,适配体与靶标结合后发生构象变化,基于这一作用机制,适配体可用于生物传感领域. 利用AuNPs优良的光学性能,适配体偶联于AuNPs表面,将其转化为可视化的比色信号;此外,将适配体进行荧光标记,设计为输出荧光信号的读取模式,由此演变出几种进行信号放大的策略,更有利于提高检测灵敏度. 本文主要综述了比色、光学和电化学适配体传感器,比色以及光学适配体传感器具有易于操作、可视化检测和价格低廉的优势,与之相比,电化学适配体传感器能够实现更高的灵敏度、可重复性和准确性. 不同类型的适配体传感器各有优势和不足,在实际的抗生素检测中需根据应用场景、检测限度等方面综合考量.
适配体生物传感技术的本质是适配体与靶标分子的结合亲和力,一般用kd表示,kd值越低,代表亲和力越高. 利用SELEX技术可以获得具有优异性能的适配体. SELEX是一种分子生物学进化技术,它可以从一个丰富度极高的核酸文库(1013—1016个)中,筛选出若干个能与靶标结合的适配体. 在适配体的筛选过程中,通过人为调控适配体的靶向性,并将其他非靶分子引入到筛选中,获得具有高特异性和高选择性的适配体. 利用“先反筛后正筛”的方法,优先洗脱非特异的DNA分子,可快速缩小适配体的筛选范围,有利于特异性适配体的富集进化. 此外,筛选过程中核酸分子的扩增步骤,可实现活性分子的指数式富集,提高其在核酸文库中的占比,更有利于其与靶标的识别与结合.
重金属、农药等有毒有害物质的排放已成为全球性的环境问题,对其进行快速、即时和可靠地检测具有重要意义. 生物传感技术作为一种高灵敏度、高特异性的分析方法,可以针对被检测对象进行不同种类的传感元件设计. 高稳定性、靶标范围广和高普适性的适配体探针成为生物传感元件的首选,利用适配体替代常规检测中的抗体,构建多种功能的适配体生物传感器. SELEX技术可实现对多种目标物(细菌、病毒、小分子、蛋白质等)的识别和筛选,以适应各种环境检测需求. 在实际检测领域中,需要一种样品预处理简单且操作方便的检测方法,而即时检测(point-of-care testing, POCT)平台可以在不依赖于专业技术人员和精密仪器的情况下,在实验室外完成对样品的采集和分析,从而达到现场快速检测的目的,降低了检测成本,在环境监测、质量监控等领域有着广泛的应用前景.
适配体传感器在抗生素检测中的研究进展
Research progress of aptasensors for antibiotic detection
-
摘要: 抗生素滥用导致的污染以及耐药菌的出现已成为全球性严重问题,对其进行快速、灵敏、即时检测在生态系统和人类健康保护方面具有重要意义. 而传统的检测方法,耗时长且需要专门的仪器设备,不能满足上述检测要求. 近年来,生物传感器作为一种有效的替代方法已广泛应用于抗生素检测. 其中,以适配体作为分子识别元件的生物传感方法由于具有稳定性高、特异性强、信号传导多样化等优势,而备受关注. 本文系统综述了现有抗生素适配体体外筛选的最新进展,并综述了基于比色、光学、电化学等信号转换模式的适配体传感器,最后对其未来在小分子污染物检测领域的应用进行了展望.Abstract: Since the environmental pollution caused by antibiotic overuse and the emergence of drug-resistant bacteria have become a serious globe problem, it is of great significance to rapidly, sensitively, and accurately detect antibiotics for the preservation of ecosystems and human health. However, the traditional detection methods often need the long detection time and specialized equipment, thus they cannot meet the above detection requirement. Recently, biosensors as useful alternatives have been widely applicated in the antibiotic detection. Among them, the aptamer-based biosensing method have attracted great interest, due to its advantages of high stability, high specificity, and diverse signaling. In this paper, we thoroughly review the in vitro selection of current antibiotic aptamers. Furthermore, we highlight the recent advances of aptasensors based on colorimetric, optical, electrochemical and other signaling modes. Additionally, we offer a forecast for the development of the field of small molecule contamination detection.
-
Key words:
- antibiotic detection /
- aptamer /
- SELEX /
- aptasensor /
- biosensing.
-

-
图 3 (A)基于间接竞争ELAA的感应传感器用于四环素检测的示意图[78];(B)基于适配体的液晶传感器用于卡那霉素检测示意图[79];(C)分裂G4 DNAzyme结合双NESA比色法检测抗生素的示意图[80]Fig. 3 (A) Schematic illustration of the aptasensor based on indirect competitive ELAAfor TC detection[78];(B) Schematic illustrationof aptamer-based LC sensor forKNM[79];(C) Schematic illustration of split G4 DNAzyme coupled with dual NESA-based colorimetric assay[80]
图 4 (A)荧光共振能量转移(FRET)适配体传感器示意图[82];(B)基于AuNPs的荧光适配体传感器示意图[84];(C)荧光响应的卡那霉素检测示意图[85];(D)脱氧核酶辅助的无标记适配体传感器示意图[86]
Figure 4. (A) Schematic diagram of a fluorescence resonance energy transfer (FRET) aptamer sensor[82]; (B) Schematic illustration of FAM-labeled aptamer and gold nanoparticles[84];(C) Schematic diagram of kanamycin detectionbasedon fluorescence-response[85];(D) Schematic diagram of the labeling free aptamer sensor with DNAzyme[86]
图 6 (A)磁性纳米球靶向性适配体传感器用于四环素检测的示意图[92];(B)基于SERS的纳米生物传感器检测土霉素的示意图[93];(C)SERS传感器用于四环素检测的目标响应释放示意图[94];(D)基于适配体识别和级联DNA网络扩增的SERS适配体传感器用于四环素检测的示意图[95]Fig. 6 (A) Schematic illustration of this proposed magnetic nanospheres-targeting aptasensor for tetracycline[92]; (B) Schematic illustration of nano-biosensor with SERS-active for the detection of OTC[93]; (C) Schematic illustration of the proposed target-responsive release SERS sensor for tetracycline (TC) detection[94]; (D) Schematic illustration of tetracycline(TC)detection based on aptamer recognition and cascade DNA network amplification Raman aptasensor[95]
图 7 (A)靶诱导和T7核酸外切酶辅助的循环扩增电化学适配体传感器示意图[100];(B)基于适配体-抗生素结合诱导的电化学信号改善的抗生素检测示意图[101];(C)基于阻抗的卡那霉素(KANA)检测适配体传感器示意图[102]Fig. 7 (A) Schematic illustration of target-induced and T7 exonuclease-aided recycling amplification homogeneous electrochemical strategy for highly sensitive detection of AMP[100]; (B) Schematic illustration of antibiotic assay based on aptamer-antibiotic binding induced electrochemical signal improvement[101];(C)Schematic illustration of aptasensor for kanamycin (KANA) detection[102]
图 8 (A)基于Y-DNA探针和靶标触发扩增策略的抗生素检测示意图[104];(B)检测卡那霉素A的信号探针位移电化学适配体传感器(SD-EAB)示意图[105];(C)利用亚甲基蓝双标记适配体或[Fe(CN)6]3-/4-进行喹诺酮类抗生素检测的电化学传感器示意图[106]Fig. 8 (A) Schematic illustration of the simultaneous detection of antibiotics based on Y-DNA probe and target-triggered amplification strategy[104]; (B) Schematic illustrations of SD-EAB for the detection ofKanamycin A[105]; (C) Schematic illustration of the electrochemical sensor for FQs detection using methylene blue (double-labelled aptamer) or [Fe(CN)6]3-/4-for the detection of ciprofloxacin[106]
图 9 (A)电化学发光传感器用于氯霉素检测的示意图[111];(B)基于分裂适配体的电化学发光传感器示意图[112];(C)电化学发光传感器组示意图及Cd2+和氨苄西林的检测策略[113]Fig. 9 (A) Schematic illustration for fabrication of the ECL aptasensor and the detection strategies for target CAP[111];(B) Schematic illustration of the split aptamer sensors[112];(C) Schematic illustration of the sensor assembly process and the detection strategies of Cd2+ and ampicillin[113]
表 1 氨基糖苷类抗生素适配体汇总表
Table 1. Summary table of aminoglycoside antibiotic aptamers
抗生素种类
Kindsof antibioti适配体名称
Name of aptamer适配体类型
Type of aptamer适配体序列
Aptamer sequences(5’-3’)解离常数
Dissociation constants (kd)参考文献
References妥布霉素
(TOB)X1 RNA CUGGUUAGUUUUGCACAGUGGTCGAUGCUAGACUUGGUUUAGGUAAUGAGUCCAAUAGUC (3 ± 1) nmol·L−1 [35] J6 RNA AGUAUAGCGAGGUUUAGCUACACUCGUGCUGAUCGUUUGGUACGGGACCUGCGUGUAGCC (2 ± 1) nmol·L−1 X1SL RNA GGGUGACUUGGUUUAGGUAAUGAGUCACCC (12 ± 5) nmol·L−1 J6SL RNA GGGACGAGGUUUAGCUACACUCGUCCC (9 ± 3) nmol·L−1 BA14-1 RNA AAGCCCCUGCAAACAUUCACGAAGUGACGUCUGAACUGCUUCGAA 500 μmol·L−1 [36] BA14-2 RNA CUACUAAUCCCAACAGCCAAAAGUGACGUCUGAACUGCUUCGAA 16 μmol·L−1 CLSE1215 DNA CCATGATTCAACTTTACTGGTCTTGTCTTGGCTAGTCGTGTGTCATTCCCGTAAGGG 0.3 μmol·L−1 [37] Ap32 DNA TAGGGAATTCGTCGACGGATCCATGGCACGTTATGCGGAGGCGGTATGATAGCGCTACTGCAGGTCGACGCATGCGCCG 56.8 nmol·L−1 [49] Ap32-2 DNA CGTCGACGGATCCATGGCACGTTATAGGTCGACG 48.4 nmol·L−1 Ap4 DNA GACTAGGCACTAGTC 42.12 nmol·L−1 [50] 卡那霉素及其衍生物 Ky2 DNA TGGGGGTTGAGGCTAAGCCGA TOB: 103 nmol·L−1 [38] KANA: 78.8 nmol·L−1 KANA-B: 84.5 nmol·L−1 卡那霉素
KANA-A3-19 DNA ATACCAGCTTATTCAATTAGCCCGGTATTGAGGTCGATCTCTTATCCTATGGCTTGTCCCCCATGGCTCGGTTATATCCAGATAGTAAGTGCAATCT 3.9 μmol·L−1 [40] K16-1 DNA CGCATACCAGCTTAGTTCAGAATTCATTGGAGCGTGGCGTGGATGCCCGATGGACCGCCCCAGGGTGCAGATAGTAAGTGCAATCTCGGC (340 ± 70) μmol·L−1 [41] K16-1c DNA CGCATACCAGCTTAGTTCAGAATTCATTGGAG (400 ± 100 )μmol·L−1 链霉素
(STR)STR1 DNA TAGGGAATTCGTCGACGGATCCGGGGTCTGGTGTTCTGCTTTGTTCTGTCGGGTCGTCTGCAGGTCGACGCATGCGCCG 199.1 nmol·L−1 [42] 8-2 DNA GCGCGCCCTAGGTACGATCGCGC 132.3 nmol·L−1 [43] A15 DNA CCCGTTTAAAGTAGTTGAGAGTATTCCGTTTCTTTGTGTC 6.07 nmol·L−1 [44] 新霉素
(NEO)Neo5 RNA GGACUGGGCGAGAAGUUUAGUCC (115 ± 25) nmol·L−1 [45] N1 RNA GGCUGCUUGUCCUUUAAUGGUCCAGUC (10 ± 2) nmol·L−1 [46] 2’OMe RNA GGCCUGGGCGAGAAGUUUAGGCC 1.24 μmol·L−1 [47] 庆大霉素
(GEN)Ap-26 DNA TAGGGAATTCGTCGACGGATCCCATCGTCTTCTTGAAGTGTGTTCTCAATAGCGTGGCTGCAGGTCGACGCATGCGCCG (14.00 ± 3.34) nmol·L−1 [48] 表 2 四环素类抗生素适配体汇总表
Table 2. Summary table of tetracycline antibiotics aptamers
抗生素种类
Kinds of antibiotic适配体名称
Name of aptamer适配体类型
Type of aptamer适配体序列
Aptamer sequences(5’-3’)解离常数
Dissociation
constants (kd)参考文献
References四环素
(TET)cb28 RNA GAGCUCAGCCUGUACUGCUGCUUAAAGCCUAAAACAUACCAGAUCGCCACCCGCGCUUUAAUCUGGAGAGGUGAAGAAUUCGACCACCUAGGCUGCACCACGG 1 μmol·L−1 [55] cb28-mini RNA GGCCUAAAACAUACCAGAUUUCGAUCUGGAGAGGUGAAGAAUUCGACCACCUAGGCCGGU 1 μmol·L−1 compact-apt RNA GAGGGAGAGGUGAAGAAUACGACCACCUAGGUACCAUUGCACUCCGGUACCUAAAACAUACCCUC 0.8 nmol·L−1 [56] RNA-apt RNA GGGCCUAAAACAUACCAGAUCGCCACCCGCGCUUUAAUCUGGAGAGGUGAAGAAUACGACCACCUAGGCUC 0.77 nmol·L−1 [62] 强力霉素
(DOX)tet-apt RNA AAAGGAGAGGUGAAGAAUACGACCACCUAGGUACCAUUGCACUACGGUACCUAAAACAUACCUUU 7 nmol·L−1 [63] 土霉素
(OTC)No.4 DNA CGTACGGAATTCGCTAGCCGACGCGCGTTGGTGGTGGATGGTGTGTTACACGTGTTGTGGATCCGAGCTCCACGTG 9.61 nmol·L−1 [57] No.5 DNA CGTACGGAATTCGCTAGCACGTTGACGCTGGTGCCCGGTTGTGGTGCGAGTGTTGTGTGGATCCGAGCTCCACGTG 12.08 nmol·L−1 No.20 DNA CGTACGGAATTCGCTAGCCGAGTTGAGCCGGGCGCGGTACGGGTACTGGTATGTGTGGGGATCCGAGCTCCACGTG 56.84 nmol·L−1 四环素类
(TCs)T7 DNA CGTACGGAATTCGCTAGCGGGCAGCGGTGGTGTGGCGGGATCTGGGGTTGTGCGGTGTGGATCCGAGCTCCACGTG 357.8 nmol·L−1 [58] T15 DNA CGTACGGAATTCGCTAGCGGAGGAACGGGTTCCAGTGTGGGGTCTATCGGGGCGTGCGGGATCCGAGCTCCACGTG 197 nmol·L−1 T19 DNA CGTACGGAATTCGCTAGCCGGGAGGGCGGGGTGTGGTATGTATTGAGCGTGGTCCGTGGGATCCGAGCTCCACGTG 424.8 nmol·L−1 T20 DNA CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGCGCGTGGATCCGAGCTCCACGTG 63.6 nmol·L−1 T22 DNA CGTACGGAATTCGCTAGCGGGCGGACGCTAGGTGGTGATGCTGTGCTACACGTGTTGTGGATCCGAGCTCCACGTG 483.5 nmol·L−1 T23 DNA CGTACGGAATTCGCTAGCGGGGGCACACATGTAGGTGCTGTCCAGGTGTGGTTGTGGTGGATCCGAGCTCCACGTG 100.6 nmol·L−1 T24 DNA CGTACGGAATTCGCTAGCGGGCGGGGGTGCTGGGGGAATGGAGTGCTGCGTGCTGCGGGGATCCGAGCTCCACGTG 70.7 nmol·L−1 A2 DNA CGGTGGTG 1.067 nmol·L−1 [59] DNA-apt DNA GTTTGTGTATTACAGTTATGTTACCCTCATTTTTCTGAAC 2.94 nmol·L−1 [64] 土霉素
(OTC)OTC3 DNA CGTACGGAATTCGCTAGCCGACGCACAGTCGCTGGTGCGTACCTGGTTGCCGTTGTGTGGATCCGAGCTCCACGTG 4.7 nmol·L−1 [65] OTC5 DNA ACGACATTCCGTTGATCTCTCCCTTTTGGGTTGGTGTCGT 147 nmol·L−1 [60] 表 3 β-内酰胺类抗生素适配体汇总表
Table 3. Summary table of β-lactam antibiotics aptamers
抗生素种类
Kinds of antibiotic适配体名称
Name of aptamer适配体类型
Type of aptamer适配体序列
Aptamer sequences (5’-3’)解离常数
Dissociation constants (kd)参考文献
References氨苄青霉素(AMP) AMP4 DNA CACGGCATGGTGGGCGTCGTG 9.4 nmol·L−1 [68] AMP17 DNA GCGGGCGGTTGTATAGCGG 13.4 nmol·L−1 AMP18 DNA GCGGGCGGTTGTATAGCGG 9.8 nmol·L−1 青霉素
(PEN)P8 DNA GGGAGGACGAAGCGGAACGAGATGTAGATGAGGCTCGATCCGAATGCGTGACGTCTATCGGAATACTCGTTTTTACGCCTCAGAAGACACGCCCGACA — [69] BBA1 DNA ATGCGGATCCCGCGCGGGTCTGAGGAGTGCGCGGTGCCAGTGAGTGCGCGAAGCTTGCGC 383.4 nmol·L−1 [70] 头孢喹肟
(CFQ)W1 DNA AGCAGCACAGAGGTCAGATGAGCAGCACAGAGGTCAGATGTGGGCGCCGACGTACTAAACCCTATGCGTGCTACCGTGAA (40.13±22.11)nmol·L−1 [71] -
[1] SHAO S C, HU Y Y, CHENG J H, et al. Research progress on distribution, migration, transformation of antibiotics and antibiotic resistance genes (ARGs) in aquatic environment[J]. Critical Reviews in Biotechnology, 2018, 38(8): 1195-1208. doi: 10.1080/07388551.2018.1471038 [2] YANG Y, YIN S, LI Y X, et al. Application of aptamers in detection and chromatographic purification of antibiotics in different matrices[J]. TrAC Trends in Analytical Chemistry, 2017, 95: 1-22. doi: 10.1016/j.trac.2017.07.023 [3] RIBEIRO Da CUNHA B, FONSECA L P, CALADO C R C. Antibiotic discovery: Where have we come from, where do we go?[J]. Antibiotics, 2019, 8(2): 45. doi: 10.3390/antibiotics8020045 [4] GRENNI P, ANCONA V, BARRA CARACCIOLO A. Ecological effects of antibiotics on natural ecosystems: A review[J]. Microchemical Journal, 2018, 136: 25-39. doi: 10.1016/j.microc.2017.02.006 [5] QIAO L N, QIAN S H, WANG Y H, et al. Carbon-dots-based lab-on-a-nanoparticle approach for the detection and differentiation of antibiotics[J]. Chemistry, 2018, 24(18): 4703-4709. doi: 10.1002/chem.201706056 [6] YI H, HUANG D L, QIN L, et al. Selective prepared carbon nanomaterials for advanced photocatalytic application in environmental pollutant treatment and hydrogen production[J]. Applied Catalysis B:Environmental, 2018, 239: 408-424. doi: 10.1016/j.apcatb.2018.07.068 [7] 姚圆, 莫测辉, 李彦文, 等. 固相萃取-高效液相色谱法分析蔬菜中四环素类抗生素[J]. 环境化学, 2010, 29(3): 536-541. YAO Y, MO C H, LI Y W, et al. Determination of tetracyclines in vegetables using solid phase extraction and hplc with fluorescence detection[J]. Environmental Chemistry, 2010, 29(3): 536-541 (in Chinese).
[8] 李振环, 朱英, 胡小键, 等. 抗生素的人体健康风险、内暴露特征及检测技术研究进展[J]. 环境化学, 2023, 42(12): 4051-4066. doi: 10.7524/j.issn.0254-6108.2022052201 LI Z H, ZHU Y, HU X J, et al. Research progress on human health risk, internal exposure characteristics and analysis technologies of antibiotics[J]. Environmental Chemistry, 2023, 42(12): 4051-4066 (in Chinese). doi: 10.7524/j.issn.0254-6108.2022052201
[9] 李颖, 黄鑫, 邹子玉, 等. 固相萃取-超高效液相色谱串联质谱同时测定鸡粪中27种抗生素[J]. 环境化学, 2023, 42(12): 4185-4194. doi: 10.7524/j.issn.0254-6108.2022060102 LI Y, HUANG X, ZOU Z Y, et al. Simultaneous determination of 27 antibiotics in chicken manure based on solid phase extraction and ultra-high performance liquid chromatographytandem mass spectrometry[J]. Environmental Chemistry, 2023, 42(12): 4185-4194 (in Chinese). doi: 10.7524/j.issn.0254-6108.2022060102
[10] 黄秋鑫, 陈琼, 雷敏, 等. 同位素内标稀释高效液相色谱-质谱法同时测定水中多种痕量抗生素[J]. 环境化学, 2016, 35(7): 1493-1499. doi: 10.7524/j.issn.0254-6108.2016.07.2015121007 HUANG Q X, CHEN Q, LEI M, et al. Simultaneous determination of trace antibiotics in surface water by isotopediluted high performance liquid chromatography-mass spectrometry[J]. Environmental Chemistry, 2016, 35(7): 1493-1499 (in Chinese). doi: 10.7524/j.issn.0254-6108.2016.07.2015121007
[11] HAN S J, ZHOU T J, YIN B J, et al. Gold nanoparticle-based colorimetric ELISA for quantification of ractopamine[J]. Microchimica Acta, 2018, 185(4): 210. doi: 10.1007/s00604-018-2736-3 [12] PAUL P, SÄNGER-VAN de GRIEND C, ADAMS E, et al. A simple, low-cost and robust capillary zone electrophoresis method with capacitively coupled contactless conductivity detection for the routine determination of four selected penicillins in money-constrained laboratories[J]. Electrophoresis, 2018, 39(20): 2521-2529. doi: 10.1002/elps.201800033 [13] 张志超, 程和发. 环境介质中喹诺酮类抗生素的前处理与检测方法研究进展[J]. 环境化学, 2019, 38(1): 1-22. doi: 10.1002/etc.4337 ZHANG Z C, CHENG H F. Recent development in sample pretreatment and detection methods for the determination of quinolones in environmental matrices[J]. Environmental Chemistry, 2019, 38(1): 1-22 (in Chinese). doi: 10.1002/etc.4337
[14] 江新泽, 常兴, 李原婷, 等. 传感器在抗生素检测中的研究进展[J]. 环境化学, 2016, 35(12): 2491-2500. doi: 10.7524/j.issn.0254-6108.2016.12.2016042502 JIANG X Z, CHANG X, LI Y T, et al. Research progress on sensors in detection of antibiotics[J]. Environmental Chemistry, 2016, 35(12): 2491-2500 (in Chinese). doi: 10.7524/j.issn.0254-6108.2016.12.2016042502
[15] YU H X, ALKHAMIS O, CANOURA J, et al. Advances and challenges in small-molecule DNA aptamer isolation, characterization, and sensor development[J]. Angewandte Chemie, 2021, 60(31): 16800-16823. doi: 10.1002/anie.202008663 [16] KHOSHBIN Z, HOUSAINDOKHT M R, VERDIAN A, et al. Simultaneous detection and determination of mercury (II) and lead (II) ions through the achievement of novel functional nucleic acid-based biosensors[J]. Biosensors and Bioelectronics, 2018, 116: 130-147. doi: 10.1016/j.bios.2018.05.051 [17] ROUSHANI M, GHANBARI K, JAFAR HOSEINI S. Designing an electrochemical aptasensor based on immobilization of the aptamer onto nanocomposite for detection of the streptomycin antibiotic[J]. Microchemical Journal, 2018, 141: 96-103. doi: 10.1016/j.microc.2018.05.016 [18] JIANG Y, SHI M L, LIU Y, et al. Aptamer/AuNP biosensor for colorimetric profiling of exosomal proteins[J]. Angewandte Chemie (International Ed. in English), 2017, 56(39): 11916-11920. doi: 10.1002/anie.201703807 [19] 段培宇, 陈寒玉, 张宝忠, 等. 动物源性食品中抗生素类污染物生物检测技术研究进展[J]. 环境化学, 2022, 41(2): 581-590. doi: 10.7524/j.issn.0254-6108.2020100704 DUAN P Y, CHEN H Y, ZHANG B Z, et al. Research progress of bioassay technology for antibiotic pollutants in animal-derived foods[J]. Environmental Chemistry, 2022, 41(2): 581-590 (in Chinese). doi: 10.7524/j.issn.0254-6108.2020100704
[20] TUERK C, GOLD L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase[J]. Science, 1990, 249(4968): 505-510. doi: 10.1126/science.2200121 [21] ELLINGTON A D, SZOSTAK J W. In vitro selection of RNA molecules that bind specific ligands[J]. Nature, 1990, 346(6287): 818-822. doi: 10.1038/346818a0 [22] LIU C B, LU C X, TANG Z G, et al. Aptamer-functionalized magnetic nanoparticles for simultaneous fluorometric determination of oxytetracycline and kanamycin[J]. Microchimica Acta, 2015, 182(15): 2567-2575. [23] GARCIA-ALVAREZ L, DAWSON S, COOKSON B, et al. Working across the veterinary and human health sectors[J]. Journal of Antimicrobial Chemotherapy, 2012, 67(suppl_1): i37-i49. [24] QIAN S W, CHANG D R, HE S S, et al. Aptamers from random sequence space: Accomplishments, gaps and future considerations[J]. Analytica Chimica Acta, 2022, 1196: 339511. doi: 10.1016/j.aca.2022.339511 [25] HERMANN T, PATEL D J. Adaptive recognition by nucleic acid aptamers[J]. Science, 2000, 287(5454): 820-825. doi: 10.1126/science.287.5454.820 [26] WARNER K D, CHEN M C, SONG W J, et al. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein[J]. Nature Structural & Molecular Biology, 2014, 21(8): 658-663. [27] SINGH V. Ultrasensitive quantum dot-coupled-surface plasmon microfluidic aptasensor array for serum insulin detection[J]. Talanta, 2020, 219: 121314. doi: 10.1016/j.talanta.2020.121314 [28] QIU W W, WANG Q X, YANO N, et al. Flexible flower-like MOF of Cu2(trans-1, 4-cyclohexanedicarboxylic acid)2 as the electroactive matrix material for label-free and highly sensitive sensing of thrombin[J]. Electrochimica Acta, 2020, 353: 136611. doi: 10.1016/j.electacta.2020.136611 [29] YUE F L, LI F L, KONG Q Q, et al. Recent advances in aptamer-based sensors for aminoglycoside antibiotics detection and their applications[J]. The Science of the Total Environment, 2021, 762: 143129. doi: 10.1016/j.scitotenv.2020.143129 [30] TANG W X, YU J, WANG Z Z, et al. Label-free potentiometric aptasensing platform for the detection of Pb2+ based on guanine quadruplex structure[J]. Analytica Chimica Acta, 2019, 1078: 53-59. doi: 10.1016/j.aca.2019.06.020 [31] MEHLHORN A, RAHIMI P, JOSEPH Y. Aptamer-based biosensors for antibiotic detection: A review[J]. Biosensors, 2018, 8(2): 54. doi: 10.3390/bios8020054 [32] PRAYLE A, WATSON A, FORTNUM H, et al. Side effects of aminoglycosides on the kidney, ear and balance in cystic fibrosis[J]. Thorax, 2010, 65(7): 654-658. doi: 10.1136/thx.2009.131532 [33] WANG X R, YANG S P, LI Y, et al. Optimization and application of parallel solid-phase extraction coupled with ultra-high performance liquid chromatography-tandem mass spectrometry for the determination of 11 aminoglycoside residues in honey and royal jelly[J]. Journal of Chromatography A, 2018, 1542: 28-36. doi: 10.1016/j.chroma.2018.02.029 [34] ZHANG Z, CAO X L, ZHANG Z P, et al. Synthesis of dummy-template molecularly imprinted polymer adsorbents for solid phase extraction of aminoglycosides antibiotics from environmental water samples[J]. Talanta, 2020, 208: 120385. doi: 10.1016/j.talanta.2019.120385 [35] WANG Y, RANDO R R. Specific binding of aminoglycoside antibiotics to RNA[J]. Chemistry & Biology, 1995, 2(5): 281-290. [36] MORSE D P. Direct selection of RNA beacon aptamers[J]. Biochemical and Biophysical Research Communications, 2007, 359(1): 94-101. doi: 10.1016/j.bbrc.2007.05.072 [37] SPIGA F M, MAIETTA P, GUIDUCCI C. More DNA-aptamers for small drugs: A capture-SELEX coupled with surface plasmon resonance and high-throughput sequencing[J]. ACS Combinatorial Science, 2015, 17(5): 326-333. doi: 10.1021/acscombsci.5b00023 [38] SONG K M, CHO M, JO H, et al. Gold nanoparticle-based colorimetric detection of kanamycin using a DNA aptamer[J]. Analytical Biochemistry, 2011, 415(2): 175-181. doi: 10.1016/j.ab.2011.04.007 [39] JALALIAN S H, KARIMABADI N, RAMEZANI M, et al. Electrochemical and optical aptamer-based sensors for detection of tetracyclines[J]. Trends in Food Science & Technology, 2018, 73: 45-57. [40] STOLTENBURG R, NIKOLAUS N, STREHLITZ B. Capture-SELEX: Selection of DNA aptamers for aminoglycoside antibiotics[J]. Journal of Analytical Methods in Chemistry, 2012, 2012: 415697. [41] SANFORD A A, RANGEL A E, FEAGIN T A, et al. RE-SELEX: Restriction enzyme-based evolution of structure-switching aptamer biosensors[J]. Chemical Science, 2021, 12(35): 11692-11702. doi: 10.1039/D1SC02715H [42] ZHOU N D, WANG J Y, ZHANG J, et al. Selection and identification of streptomycin-specific single-stranded DNA aptamers and the application in the detection of streptomycin in honey[J]. Talanta, 2013, 108: 109-116. doi: 10.1016/j.talanta.2013.01.064 [43] SOHEILI V, TAGHDISI S M, KHAYYAT M H, et al. Colorimetric and ratiometric aggregation assay for streptomycin using gold nanoparticles and a new and highly specific aptamer[J]. Microchimica Acta, 2016, 183(5): 1687-1697. doi: 10.1007/s00604-016-1798-3 [44] LIU Z C, ZHANG Y F, XIE Y, et al. An aptamer-based colorimetric sensor for streptomycin and its application in food inspection[J]. Chemical Research in Chinese Universities, 2017, 33(5): 714-720. doi: 10.1007/s40242-017-7029-6 [45] WALLIS M G, von AHSEN U, SCHROEDER R, et al. A novel RNA motif for neomycin recognition[J]. Chemistry & Biology, 1995, 2(8): 543-552. [46] WEIGAND J E, SANCHEZ M, GUNNESCH E B, et al. Screening for engineered neomycin riboswitches that control translation initiation[J]. RNA, 2008, 14(1): 89-97. doi: 10.1261/rna.772408 [47] DE-LOS-SANTOS-ÁLVAREZ N, LOBO-CASTAÑÓN M J, MIRANDA-ORDIERES A J, et al. SPR sensing of small molecules with modified RNA aptamers: Detection of neomycin B[J]. Biosensors and Bioelectronics, 2009, 24(8): 2547-2553. doi: 10.1016/j.bios.2009.01.011 [48] 巫朦朦, 韩旭艳, 蔡蓉凤, 等. 庆大霉素特异性单链DNA适配体的筛选、表征和应用[J]. 中国科学:生命科学, 2019, 49(5): 637-648. doi: 10.1360/N052018-00238 WU M M, HAN X Y, CAI R F, et al. Selection, characterization and application of gentamicin-specific single-stranded DNA Aptamers[J]. Scientia Sinica (Vitae), 2019, 49(5): 637-648 (in Chinese). doi: 10.1360/N052018-00238
[49] HAN X Y, ZHANG Y H, NIE J J, et al. Gold nanoparticle based photometric determination of tobramycin by using new specific DNA aptamers[J]. Microchimica Acta, 2018, 185(1): 4. doi: 10.1007/s00604-017-2568-6 [50] NIE J J, YUAN L Y, JIN K, et al. Electrochemical detection of tobramycin based on enzymes-assisted dual signal amplification by using a novel truncated aptamer with high affinity[J]. Biosensors and Bioelectronics, 2018, 122: 254-262. doi: 10.1016/j.bios.2018.09.072 [51] TAGHDISI S M, DANESH N M, RAMEZANI M, et al. A novel M-shape electrochemical aptasensor for ultrasensitive detection of tetracyclines[J]. Biosensors and Bioelectronics, 2016, 85: 509-514. doi: 10.1016/j.bios.2016.05.048 [52] ZHAO H M, GAO S, LIU M, et al. Fluorescent assay for oxytetracycline based on a long-chain aptamer assembled onto reduced graphene oxide[J]. Microchimica Acta, 2013, 180(9): 829-835. [53] LIU Y, KONG J J, YUAN J L, et al. Enhanced photocatalytic activity over flower-like sphere Ag/Ag2CO3/BiVO4 plasmonic heterojunction photocatalyst for tetracycline degradation[J]. Chemical Engineering Journal, 2018, 331: 242-254. doi: 10.1016/j.cej.2017.08.114 [54] ZAHRA Q U A, LUO Z F, ALI R, et al. Advances in gold nanoparticles-based colorimetric aptasensors for the detection of antibiotics: An overview of the past decade[J]. Nanomaterials, 2021, 11(4): 840. doi: 10.3390/nano11040840 [55] BERENS C, THAIN A, SCHROEDER R. A tetracycline-binding RNA aptamer[J]. Bioorganic & Medicinal Chemistry, 2001, 9(10): 2549-2556. [56] XIAO H, EDWARDS T E, FERRÉ-D'AMARÉ A R. Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch[J]. Chemistry & Biology, 2008, 15(10): 1125-1137. [57] NIAZI J H, LEE S J, KIM Y S, et al. ssDNA aptamers that selectively bind oxytetracycline[J]. Bioorganic & Medicinal Chemistry, 2008, 16(3): 1254-1261. [58] NIAZI J H, LEE S J, GU M B. Single-stranded DNA aptamers specific for antibiotics tetracyclines[J]. Bioorganic & Medicinal Chemistry, 2008, 16(15): 7245-7253. [59] KWON Y S, AHMAD RASTON N H, GU M B. An ultra-sensitive colorimetric detection of tetracyclines using the shortest aptamer with highly enhanced affinity[J]. Chemical Communications, 2014, 50(1): 40-42. doi: 10.1039/C3CC47108J [60] ZHAO Y C, ONG S, CHEN Y J, et al. Label-free and dye-free fluorescent sensing of tetracyclines using a capture-selected DNA aptamer[J]. Analytical Chemistry, 2022, 94(28): 10175-10182. doi: 10.1021/acs.analchem.2c01561 [61] JEONG S, PAENG I R. Sensitivity and selectivity on aptamer-based assay: The determination of tetracycline residue in bovine milk[J]. The Scientific World Journal, 2012, 2012: 159456. [62] MÜLLER M, WEIGAND J E, WEICHENRIEDER O, et al. Thermodynamic characterization of an engineered tetracycline-binding riboswitch[J]. Nucleic Acids Research, 2006, 34(9): 2607-2617. doi: 10.1093/nar/gkl347 [63] TICKNER Z J, ZHONG G C, SHEPTACK K R, et al. Selection of high-affinity RNA aptamers that distinguish between doxycycline and tetracycline[J]. Biochemistry, 2020, 59(37): 3473-3486. doi: 10.1021/acs.biochem.0c00586 [64] WANG S, LIU J H, YONG W, et al. A direct competitive assay-based aptasensor for sensitive determination of tetracycline residue in Honey[J]. Talanta, 2015, 131: 562-569. doi: 10.1016/j.talanta.2014.08.028 [65] KIM C H, LEE L P, MIN J R, et al. An indirect competitive assay-based aptasensor for detection of oxytetracycline in milk[J]. Biosensors and Bioelectronics, 2014, 51: 426-430. doi: 10.1016/j.bios.2013.08.003 [66] KELLMANN M, MUENSTER H, ZOMER P, et al. Full scan MS in comprehensive qualitative and quantitative residue analysis in food and feed matrices: How much resolving power is required?[J]. Journal of the American Society for Mass Spectrometry, 2009, 20(8): 1464-1476. doi: 10.1016/j.jasms.2009.05.010 [67] ABEDALWAFA M A, LI Y, NI C F, et al. Colorimetric sensor arrays for the detection and identification of antibiotics[J]. Analytical Methods, 2019, 11(22): 2836-2854. doi: 10.1039/C9AY00371A [68] SONG K M, JEONG E, JEON W, et al. Aptasensor for ampicillin using gold nanoparticle based dual fluorescence-colorimetric methods[J]. Analytical and Bioanalytical Chemistry, 2012, 402(6): 2153-2161. doi: 10.1007/s00216-011-5662-3 [69] PANIEL N, ISTAMBOULIÉ G, TRIKI A, et al. Selection of DNA aptamers against penicillin G using Capture-SELEX for the development of an impedimetric sensor[J]. Talanta, 2017, 162: 232-240. doi: 10.1016/j.talanta.2016.09.058 [70] LEE A Y, HA N R, JUNG I P, et al. Development of a ssDNA aptamer for detection of residual benzylpenicillin[J]. Analytical Biochemistry, 2017, 531: 1-7. doi: 10.1016/j.ab.2017.05.013 [71] WANG L H, WANG C C, LI H. Selection of DNA aptamers and establishment of an effective aptasensor for highly sensitive detection of cefquinome residues in milk[J]. The Analyst, 2018, 143(13): 3202-3208. doi: 10.1039/C8AN00709H [72] XU Y Y, HAN T, LI X Q, et al. Colorimetric detection of kanamycin based on analyte-protected silver nanoparticles and aptamer-selective sensing mechanism[J]. Analytica Chimica Acta, 2015, 891: 298-303. doi: 10.1016/j.aca.2015.08.013 [73] DENG C Y, LIU H, ZHANG M M, et al. Light-up nonthiolated aptasensor for low-mass, soluble amyloid-β40 oligomers at high salt concentrations[J]. Analytical Chemistry, 2018, 90(3): 1710-1717. doi: 10.1021/acs.analchem.7b03468 [74] EPANCHINTSEVA A, VOROBJEV P, PYSHNYI D, et al. Fast and strong adsorption of native oligonucleotides on citrate-coated gold nanoparticles[J]. Langmuir:the ACS Journal of Surfaces and Colloids, 2018, 34(1): 164-172. doi: 10.1021/acs.langmuir.7b02529 [75] SAHA K, AGASTI S S, KIM C, et al. Gold nanoparticles in chemical and biological sensing[J]. Chemical Reviews, 2012, 112(5): 2739-2779. doi: 10.1021/cr2001178 [76] ZHOU N D, LUO J B, ZHANG J, et al. A label-free electrochemical aptasensor for the detection of kanamycin in milk[J]. Analytical Methods, 2015, 7(5): 1991-1996. doi: 10.1039/C4AY02710H [77] NIU S C, LV Z Z, LIU J C, et al. Colorimetric aptasensor using unmodified gold nanoparticles for homogeneous multiplex detection[J]. PLoS One, 2014, 9(10): e109263. doi: 10.1371/journal.pone.0109263 [78] WANG S, YONG W, LIU J H, et al. Development of an indirect competitive assay-based aptasensor for highly sensitive detection of tetracycline residue in honey[J]. Biosensors and Bioelectronics, 2014, 57: 192-198. doi: 10.1016/j.bios.2014.02.032 [79] WANG Y, WANG B, SHEN J, et al. Aptamer based bare eye detection of kanamycin by using a liquid crystal film on a glass support[J]. Microchimica Acta, 2017, 184(10): 3765-3771. doi: 10.1007/s00604-017-2405-y [80] CUI X J, LI R G, LIU X F, et al. Low-background and visual detection of antibiotic based on target-activated colorimetric split peroxidase DNAzyme coupled with dual nicking enzyme signal amplification[J]. Analytica Chimica Acta, 2018, 997: 1-8. doi: 10.1016/j.aca.2017.10.009 [81] 余杰, 张宴, 任洪强. 基于共振能量转移的生物传感器用于环境检测的研究进展[J]. 环境化学, 2023, 42(12): 4171-4184. doi: 10.7524/j.issn.0254-6108.2022052902 YU J, ZHANG Y, REN H Q. Research progress of biosensors based on resonance energy transfer in the field of environmental pollutant detection[J]. Environmental Chemistry, 2023, 42(12): 4171-4184 (in Chinese). doi: 10.7524/j.issn.0254-6108.2022052902
[82] LI H, SUN D E, LIU Y J, et al. An ultrasensitive homogeneous aptasensor for kanamycin based on upconversion fluorescence resonance energy transfer[J]. Biosensors and Bioelectronics, 2014, 55: 149-156. doi: 10.1016/j.bios.2013.11.079 [83] LIAO Q G, WEI B H, LUO L G. Aptamer based fluorometric determination of kanamycin using double-stranded DNA and carbon nanotubes[J]. Microchimica Acta, 2017, 184(2): 627-632. doi: 10.1007/s00604-016-2050-x [84] CHEN J, LI Z H, GE J, et al. An aptamer-based signal-on bio-assay for sensitive and selective detection of Kanamycin A by using gold nanoparticles[J]. Talanta, 2015, 139: 226-232. doi: 10.1016/j.talanta.2015.02.036 [85] BELAL A S F, ISMAIL A, ELNAGGAR M M, et al. Click chemistry inspired copper sulphide nanoparticle-based fluorescence assay of kanamycin using DNA aptamer[J]. Spectrochimica Acta Part A:Molecular and Biomolecular Spectroscopy, 2018, 205: 48-54. doi: 10.1016/j.saa.2018.07.011 [86] ZHOU W J, XU L, JIANG B Y. Target-initiated autonomous synthesis of metal-ion dependent DNAzymes for label-free and amplified fluorescence detection of kanamycin in milk samples[J]. Analytica Chimica Acta, 2021, 1148: 238195. doi: 10.1016/j.aca.2020.12.070 [87] RAMEZANI M, DANESH N M, LAVAEE P, et al. A selective and sensitive fluorescent aptasensor for detection of kanamycin based on catalytic recycling activity of exonuclease III and gold nanoparticles[J]. Sensors and Actuators B:Chemical, 2016, 222: 1-7. doi: 10.1016/j.snb.2015.08.024 [88] LEUNG K H, HE H Z, CHAN D S H, et al. An oligonucleotide-based switch-on luminescent probe for the detection of kanamycin in aqueous solution[J]. Sensors and Actuators B:Chemical, 2013, 177: 487-492. doi: 10.1016/j.snb.2012.11.053 [89] HAO L L, GU H J, DUAN N, et al. A chemiluminescent aptasensor for simultaneous detection of three antibiotics in milk[J]. Analytical Methods, 2016, 8(44): 7929-7936. doi: 10.1039/C6AY02304E [90] YANG K, HU Y J, DONG N. A novel biosensor based on competitive SERS immunoassay and magnetic separation for accurate and sensitive detection of chloramphenicol[J]. Biosensors and Bioelectronics, 2016, 80: 373-377. doi: 10.1016/j.bios.2016.01.064 [91] GE L, LI H N, DU X J, et al. Facile one-pot synthesis of visible light-responsive BiPO4/nitrogen doped graphene hydrogel for fabricating label-free photoelectrochemical tetracycline aptasensor[J]. Biosensors and Bioelectronics, 2018, 111: 131-137. doi: 10.1016/j.bios.2018.04.008 [92] LI H H, CHEN Q S, HASSAN M M, et al. A magnetite/PMAA nanospheres-targeting SERS aptasensor for tetracycline sensing using mercapto molecules embedded core/shell nanoparticles for signal amplification[J]. Biosensors and Bioelectronics, 2017, 92: 192-199. doi: 10.1016/j.bios.2017.02.009 [93] MENG F W, MA X Y, DUAN N, et al. Ultrasensitive SERS aptasensor for the detection of oxytetracycline based on a gold-enhanced nano-assembly[J]. Talanta, 2017, 165: 412-418. doi: 10.1016/j.talanta.2016.12.088 [94] LI H H, GENG W H, HARUNA S A, et al. A target-responsive release SERS sensor for sensitive detection of tetracycline using aptamer-gated HP-UiO-66-NH2 nanochannel strategy[J]. Analytica Chimica Acta, 2022, 1220: 339999. doi: 10.1016/j.aca.2022.339999 [95] LV Y, QI S, KHAN I M, et al. Concatenated dynamic DNA network modulated SERS aptasensor based on gold-magnetic nanochains and Au@Ag nanoparticles for enzyme-free amplification analysis of tetracycline[J]. Analytica Chimica Acta, 2023, 1270: 341238. doi: 10.1016/j.aca.2023.341238 [96] PFEIFFER F, MAYER G. Selection and biosensor application of aptamers for small molecules[J]. Frontiers in Chemistry, 2016, 4: 25. [97] SONG S P, WANG L H, LI J, et al. Aptamer-based biosensors[J]. TrAC Trends in Analytical Chemistry, 2008, 27(2): 108-117. doi: 10.1016/j.trac.2007.12.004 [98] DAPRÀ J, LAURIDSEN L H, NIELSEN A T, et al. Comparative study on aptamers as recognition elements for antibiotics in a label-free all-polymer biosensor[J]. Biosensors and Bioelectronics, 2013, 43: 315-320. doi: 10.1016/j.bios.2012.12.058 [99] ROSATI G, DAPRÀ J, CHERRÉ S, et al. Performance improvement by layout designs of conductive polymer microelectrode based impedimetric biosensors[J]. Electroanalysis, 2014, 26(6): 1400-1408. doi: 10.1002/elan.201400062 [100] WANG X Z, DONG S S, GAI P P, et al. Highly sensitive homogeneous electrochemical aptasensor for antibiotic residues detection based on dual recycling amplification strategy[J]. Biosensors and Bioelectronics, 2016, 82: 49-54. doi: 10.1016/j.bios.2016.03.055 [101] WANG J, MA K, YIN H S, et al. Aptamer based voltammetric determination of ampicillin using a single-stranded DNA binding protein and DNA functionalized gold nanoparticles[J]. Microchimica Acta, 2018, 185(1): 68. doi: 10.1007/s00604-017-2566-8 [102] SHARMA A, ISTAMBOULIE G, HAYAT A, et al. Disposable and portable aptamer functionalized impedimetric sensor for detection of kanamycin residue in milk sample[J]. Sensors and Actuators B:Chemical, 2017, 245: 507-515. doi: 10.1016/j.snb.2017.02.002 [103] SUN X, LI F L, SHEN G H, et al. Aptasensor based on the synergistic contributions of chitosan-gold nanoparticles, graphene-gold nanoparticles and multi-walled carbon nanotubes-cobalt phthalocyanine nanocomposites for kanamycin detection[J]. Analyst, 2014, 139(1): 299-308. doi: 10.1039/C3AN01840G [104] CHEN M, GAN N, LI T H, et al. An electrochemical aptasensor for multiplex antibiotics detection using Y-shaped DNA-based metal ions encoded probes with NMOF substrate and CSRP target-triggered amplification strategy[J]. Analytica Chimica Acta, 2017, 968: 30-39. doi: 10.1016/j.aca.2017.03.024 [105] LIU R, YANG Z H, GUO Q, et al. Signaling-probe displacement electrochemical aptamer-based sensor (SD-EAB) for detection of nanomolar kanamycin A[J]. Electrochimica Acta, 2015, 182: 516-523. doi: 10.1016/j.electacta.2015.09.140 [106] TAGHDISI HEIDARIAN S M, TAVANAEE SANI A, DANESH N M, et al. A novel electrochemical approach for the ultrasensitive detection of fluoroquinolones based on a double-labelled aptamer to surpass complementary strands of aptamer lying flat[J]. Sensors and Actuators B:Chemical, 2021, 334: 129632. doi: 10.1016/j.snb.2021.129632 [107] ALTHOMALI R H, HAMOUD ALSHAHRANI S, QASIM ALMAJIDI Y, et al. Current trends in nanomaterials-based electrochemiluminescence aptasensors for the determination of antibiotic residues in foodstuffs: A comprehensive review[J]. Critical Reviews in Analytical Chemistry, 2023(22): 1-17. [108] SHEN Y Z, GAO X, LU H J, et al. Electrochemiluminescence-based innovative sensors for monitoring the residual levels of heavy metal ions in environment-related matrices[J]. Coordination Chemistry Reviews, 2023, 476: 214927. doi: 10.1016/j.ccr.2022.214927 [109] ZHONG X, LI X, ZHUO Y, et al. Synthesizing anode electrochemiluminescent self-catalyzed carbon dots-based nanocomposites and its application in sensitive ECL biosensor for microRNA detection[J]. Sensors and Actuators B:Chemical, 2020, 305: 127490. doi: 10.1016/j.snb.2019.127490 [110] ZHAO Y, WANG R Z, WANG Y H, et al. Dual-channel molecularly imprinted sensor based on dual-potential electrochemiluminescence of Zn-MOFs for double detection of trace chloramphenicol[J]. Food Chemistry, 2023, 413: 135627. doi: 10.1016/j.foodchem.2023.135627 [111] WEN J, JIANG D, SHAN X L, et al. Ternary electrochemiluminescence biosensor based on black phosphorus quantum dots doped perylene derivative and metal organic frameworks as a coreaction accelerator for the detection of chloramphenicol[J]. Microchemical Journal, 2022, 172: 106927. doi: 10.1016/j.microc.2021.106927 [112] ZHANG X Y, DU Y, LIU X J, et al. Enhanced anode electrochemiluminescence in split aptamer sensor for kanamycin trace monitoring[J]. Food Chemistry, 2023, 420: 136083. doi: 10.1016/j.foodchem.2023.136083 [113] ZHAI H G, WANG Y, GENG L J, et al. Bipotential-resolved electrochemiluminescence biosensor based on Bi2S3@Au nanoflowers for simultaneous detection of Cd(II) and ampicillin in aquatic products[J]. Food Chemistry, 2023, 414: 135708. doi: 10.1016/j.foodchem.2023.135708 [114] BAI X J, HOU H, ZHANG B L, et al. Label-free detection of kanamycin using aptamer-based cantilever array sensor[J]. Biosensors and Bioelectronics, 2014, 56: 112-116. doi: 10.1016/j.bios.2013.12.068 -



 下载:
下载: